Contributing Factors for Mutagenic DNA Lesion Bypass by DNA Polymerase Eta (polη)
Abstract
:1. Introduction
2. Overall and Catalytic Domain Structures of polη
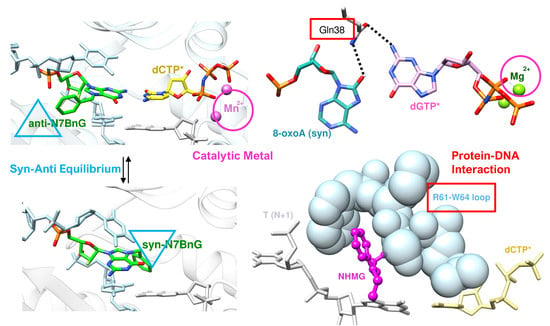
3. Catalytic Metal Cofactors—Effect of Displacement of Mg2+ by Mn2+
4. Syn–Anti Conformational Change
5. Enol–Keto Tautomerization and polη Residues
6. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Drablos, F.; Feyzi, E.; Aas, P.A.; Vaagbo, C.B.; Kavli, B.; Bratlie, M.S.; Pena-Diaz, J.; Otterlei, M.; Slupphaug, G.; Krokan, H.E. Alkylation damage in DNA and RNA—Repair mechanisms and medical significance. DNA Repair 2004, 3, 1389–1407. [Google Scholar] [CrossRef] [PubMed]
- Eadie, J.S.; Conrad, M.; Toorchen, D.; Topal, M.D. Mechanism of mutagenesis by O6-methylguanine. Nature 1984, 308, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Monti, P.; Traverso, I.; Casolari, L.; Menichini, P.; Inga, A.; Ottaggio, L.; Russo, D.; Iyer, P.; Gold, B.; Fronza, G. Mutagenicity of N3-methyladenine: A multi-translesion polymerase affair. Mutat. Res. 2010, 683, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Gates, K.S.; Nooner, T.; Dutta, S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol. 2004, 17, 839–856. [Google Scholar] [CrossRef]
- Hemnani, T.; Parihar, M.S. Reactive oxygen species and oxidative DNA damage. Indian J. Physiol. Pharmacol. 1998, 42, 440–452. [Google Scholar] [PubMed]
- Ames, B.N. Endogenous oxidative DNA damage, aging, and cancer. Free Radic. Res. Commun. 1989, 7, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Talhaoui, I.; Couve, S.; Ishchenko, A.A.; Kunz, C.; Schar, P.; Saparbaev, M. 7,8-Dihydro-8-oxoadenine, a highly mutagenic adduct, is repaired by Escherichia coli and human mismatch-specific uracil/thymine-DNA glycosylases. Nucleic Acids Res. 2013, 41, 912–923. [Google Scholar] [CrossRef]
- Chen, W.; Balakrishnan, K.; Kuang, Y.; Han, Y.; Fu, M.; Gandhi, V.; Peng, X. Reactive oxygen species (ROS) inducible DNA cross-linking agents and their effect on cancer cells and normal lymphocytes. J. Med. Chem. 2014, 57, 4498–4510. [Google Scholar] [CrossRef]
- Kow, Y.W. Repair of deaminated bases in DNA. Free Radic. Biol. Med. 2002, 33, 886–893. [Google Scholar] [CrossRef]
- Caulfield, J.L.; Wishnok, J.S.; Tannenbaum, S.R. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem. 1998, 273, 12689–12695. [Google Scholar] [CrossRef] [Green Version]
- Wink, D.A.; Kasprzak, K.S.; Maragos, C.M.; Elespuru, R.K.; Misra, M.; Dunams, T.M.; Cebula, T.A.; Koch, W.H.; Andrews, A.W.; Allen, J.S.; et al. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science 1991, 254, 1001–1003. [Google Scholar] [CrossRef] [PubMed]
- Surova, O.; Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 2013, 32, 3789–3797. [Google Scholar] [CrossRef] [PubMed]
- Norbury, C.J.; Zhivotovsky, B. DNA damage-induced apoptosis. Oncogene 2004, 23, 2797–2808. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Rayala, N.K.; Lee, S. Translesion synthesis of the major nitrogen mustard-induced DNA lesion by human DNA polymerase eta. Biochem. J. 2020, 477, 4543–4558. [Google Scholar] [CrossRef]
- Gregory, M.T.; Park, G.Y.; Johnstone, T.C.; Lee, Y.S.; Yang, W.; Lippard, S.J. Structural and mechanistic studies of polymerase eta bypass of phenanthriplatin DNA damage. Proc. Natl. Acad. Sci. USA 2014, 111, 9133–9138. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hazra, T.K.; Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008, 18, 27–47. [Google Scholar] [CrossRef]
- Kusakabe, M.; Onishi, Y.; Tada, H.; Kurihara, F.; Kusao, K.; Furukawa, M.; Iwai, S.; Yokoi, M.; Sakai, W.; Sugasawa, K. Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 2019, 41, 2. [Google Scholar] [CrossRef]
- Pecina-Slaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Knobel, P.A.; Marti, T.M. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef]
- Yang, W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase eta. Biochemistry 2014, 53, 2793–2803. [Google Scholar] [CrossRef]
- Kraemer, K.H.; Slor, H. Xeroderma pigmentosum. Clin. Dermatol. 1985, 3, 33–69. [Google Scholar] [CrossRef]
- Setlow, R.B.; Regan, J.D.; German, J.; Carrier, W.L. Evidence that xeroderma pigmentosum cells do not perform the first step in the repair of ultraviolet damage to their DNA. Proc. Natl. Acad. Sci. USA 1969, 64, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, J.E. Defective repair replication of DNA in xeroderma pigmentosum. Nature 1968, 218, 652–656. [Google Scholar] [CrossRef]
- Epstein, J.H.; Fukuyama, K.; Reed, W.B.; Epstein, W.L. Defect in DNA synthesis in skin of patients with xeroderma pigmentosum demonstrated in vivo. Science 1970, 168, 1477–1478. [Google Scholar] [CrossRef]
- Johnson, R.E.; Prakash, S.; Prakash, L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 1999, 283, 1001–1004. [Google Scholar] [CrossRef]
- Masutani, C.; Kusumoto, R.; Yamada, A.; Dohmae, N.; Yokoi, M.; Yuasa, M.; Araki, M.; Iwai, S.; Takio, K.; Hanaoka, F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 1999, 399, 700–704. [Google Scholar] [CrossRef]
- Matsuda, T.; Bebenek, K.; Masutani, C.; Hanaoka, F.; Kunkel, T.A. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature 2000, 404, 1011–1013. [Google Scholar] [CrossRef] [PubMed]
- Masutani, C.; Araki, M.; Yamada, A.; Kusumoto, R.; Nogimori, T.; Maekawa, T.; Iwai, S.; Hanaoka, F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999, 18, 3491–3501. [Google Scholar] [CrossRef] [PubMed]
- Biertumpfel, C.; Zhao, Y.; Kondo, Y.; Ramon-Maiques, S.; Gregory, M.; Lee, J.Y.; Masutani, C.; Lehmann, A.R.; Hanaoka, F.; Yang, W. Structure and mechanism of human DNA polymerase eta. Nature 2010, 465, 1044–1048. [Google Scholar] [CrossRef]
- Su, Y.; Patra, A.; Harp, J.M.; Egli, M.; Guengerich, F.P. Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2′-deoxyguanosine. J. Biol. Chem. 2015, 290, 15921–15933. [Google Scholar] [CrossRef]
- Koag, M.C.; Jung, H.; Lee, S. Mutagenic Replication of the Major Oxidative Adenine Lesion 7,8-Dihydro-8-oxoadenine by Human DNA Polymerases. J. Am. Chem. Soc. 2019, 141, 4584–4596. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Jung, H.; Lee, S. Mutagenesis mechanism of the major oxidative adenine lesion 7,8-dihydro-8-oxoadenine. Nucleic Acids Res. 2020, 48, 5119–5134. [Google Scholar] [CrossRef]
- Patra, A.; Zhang, Q.; Lei, L.; Su, Y.; Egli, M.; Guengerich, F.P. Structural and kinetic analysis of nucleoside triphosphate incorporation opposite an abasic site by human translesion DNA polymerase eta. J. Biol. Chem. 2015, 290, 8028–8038. [Google Scholar] [CrossRef]
- Zhao, Y.; Biertumpfel, C.; Gregory, M.T.; Hua, Y.J.; Hanaoka, F.; Yang, W. Structural basis of human DNA polymerase eta-mediated chemoresistance to cisplatin. Proc. Natl. Acad. Sci. USA 2012, 109, 7269–7274. [Google Scholar] [CrossRef]
- Koag, M.C.; Jung, H.; Kou, Y.; Lee, S. Bypass of the Major Alkylative DNA Lesion by Human DNA Polymerase eta. Molecules 2019, 24, 3928. [Google Scholar] [CrossRef]
- Jung, H.; Rayala, N.K.; Lee, S. Effects of N7-Alkylguanine Conformation and Metal Cofactors on the Translesion Synthesis by Human DNA Polymerase eta. Chem. Res. Toxicol. 2022, 35, 512–521. [Google Scholar] [CrossRef]
- Jung, H.; Hawkins, M.; Lee, S. Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase. Biochem. J. 2020, 477, 4797–4810. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Cleaver, J.E.; Hatahet, Z.; Honkanen, R.E.; Chang, J.Y.; Yen, Y.; Chou, K.M. Human DNA polymerase eta activity and translocation is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 16578–16583. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; You, C.; Wang, Y. The Functions of Serine 687 Phosphorylation of Human DNA Polymerase eta in UV Damage Tolerance. Mol. Cell. Proteom. 2016, 15, 1913–1920. [Google Scholar] [CrossRef]
- Kannouche, P.L.; Wing, J.; Lehmann, A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 2004, 14, 491–500. [Google Scholar] [CrossRef]
- Bienko, M.; Green, C.M.; Sabbioneda, S.; Crosetto, N.; Matic, I.; Hibbert, R.G.; Begovic, T.; Niimi, A.; Mann, M.; Lehmann, A.R.; et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol. Cell 2010, 37, 396–407. [Google Scholar] [CrossRef]
- Bomar, M.G.; Pai, M.T.; Tzeng, S.R.; Li, S.S.; Zhou, P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep. 2007, 8, 247–251. [Google Scholar] [CrossRef]
- Pozhidaeva, A.; Pustovalova, Y.; D’Souza, S.; Bezsonova, I.; Walker, G.C.; Korzhnev, D.M. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase eta. Biochemistry 2012, 51, 5506–5520. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Ouzon-Shubeita, H.; Baker, M.; Koag, M.C.; Lee, S. Structural basis for the bypass of the major oxaliplatin-DNA adducts by human DNA polymerase eta. Biochem. J. 2019, 476, 747–758. [Google Scholar] [CrossRef]
- Patra, A.; Zhang, Q.; Guengerich, F.P.; Egli, M. Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2′-deoxyguanosine by Human DNA Polymerase eta. J. Biol. Chem. 2016, 291, 24304–24313. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Gregory, M.T.; Biertumpfel, C.; Hua, Y.J.; Hanaoka, F.; Yang, W. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta. Proc. Natl. Acad. Sci. USA 2013, 110, 8146–8151. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Nagy, L.D.; Zhang, Q.; Su, Y.; Muller, L.; Guengerich, F.P.; Egli, M. Kinetics, structure, and mechanism of 8-Oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase eta. J. Biol. Chem. 2014, 289, 16867–16882. [Google Scholar] [CrossRef] [PubMed]
- Weng, P.J.; Gao, Y.; Gregory, M.T.; Wang, P.; Wang, Y.; Yang, W. Bypassing a 8,5′-cyclo-2′-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution. Proc. Natl. Acad. Sci. USA 2018, 115, 10660–10665. [Google Scholar] [CrossRef]
- Patra, A.; Banerjee, S.; Johnson Salyard, T.L.; Malik, C.K.; Christov, P.P.; Rizzo, C.J.; Stone, M.P.; Egli, M. Structural Basis for Error-Free Bypass of the 5-N-Methylformamidopyrimidine-dG Lesion by Human DNA Polymerase eta and Sulfolobus solfataricus P2 Polymerase IV. J. Am. Chem. Soc. 2015, 137, 7011–7014. [Google Scholar] [CrossRef]
- Su, Y.; Egli, M.; Guengerich, F.P. Mechanism of Ribonucleotide Incorporation by Human DNA Polymerase eta. J. Biol. Chem. 2016, 291, 3747–3756. [Google Scholar] [CrossRef] [PubMed]
- Beese, L.S.; Steitz, T.A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991, 10, 25–33. [Google Scholar] [CrossRef]
- Kaushik, N.; Pandey, V.N.; Modak, M.J. Significance of the O-helix residues of Escherichia coli DNA polymerase I in DNA synthesis: Dynamics of the dNTP binding pocket. Biochemistry 1996, 35, 7256–7266. [Google Scholar] [CrossRef]
- Doublie, S.; Tabor, S.; Long, A.M.; Richardson, C.C.; Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 1998, 391, 251–258. [Google Scholar] [CrossRef]
- Ling, H.; Boudsocq, F.; Woodgate, R.; Yang, W. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell 2001, 107, 91–102. [Google Scholar] [CrossRef]
- Jung, H.; Lee, S. Promutagenic bypass of 7,8-dihydro-8-oxoadenine by translesion synthesis DNA polymerase Dpo4. Biochem. J. 2020, 477, 2859–2871. [Google Scholar] [CrossRef]
- Franklin, M.C.; Wang, J.; Steitz, T.A. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 2001, 105, 657–667. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Prasad, R.; Wilson, S.H.; Kraut, J.; Pelletier, H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry 1997, 36, 11205–11215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Pence, M.G.; Christov, P.P.; Wawrzak, Z.; Choi, J.Y.; Rizzo, C.J.; Egli, M.; Guengerich, F.P. Basis of miscoding of the DNA adduct N2,3-ethenoguanine by human Y-family DNA polymerases. J. Biol. Chem. 2012, 287, 35516–35526. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Zhao, Y.; Yamagata, Y.; Hua, Y.J.; Yang, W. Watching DNA polymerase eta make a phosphodiester bond. Nature 2012, 487, 196–201. [Google Scholar] [CrossRef] [PubMed]
- El-Deiry, W.S.; Downey, K.M.; So, A.G. Molecular mechanisms of manganese mutagenesis. Proc. Natl. Acad. Sci. USA 1984, 81, 7378–7382. [Google Scholar] [CrossRef]
- Beckman, R.A.; Mildvan, A.S.; Loeb, L.A. On the fidelity of DNA replication: Manganese mutagenesis in vitro. Biochemistry 1985, 24, 5810–5817. [Google Scholar] [CrossRef]
- Miyaki, M.; Murata, I.; Osabe, M.; Ono, T. Effect of metal cations on misincorporation by E. coli DNA polymerases. Biochem. Biophys. Res. Commun. 1977, 77, 854–860. [Google Scholar] [CrossRef]
- Sirover, M.A.; Loeb, L.A. On the fidelity of DNA replication. Effect of metal activators during synthesis with avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 1977, 252, 3605–3610. [Google Scholar] [CrossRef]
- Hays, H.; Berdis, A.J. Manganese substantially alters the dynamics of translesion DNA synthesis. Biochemistry 2002, 41, 4771–4778. [Google Scholar] [CrossRef]
- Tabor, S.; Richardson, C.C. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. USA 1989, 86, 4076–4080. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, H.; Sawaya, M.R.; Wolfle, W.; Wilson, S.H.; Kraut, J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry 1996, 35, 12762–12777. [Google Scholar] [CrossRef]
- Villani, G.; Tanguy Le Gac, N.; Wasungu, L.; Burnouf, D.; Fuchs, R.P.; Boehmer, P.E. Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase. Nucleic Acids Res. 2002, 30, 3323–3332. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, O.; Ruiz, J.F.; Lain de Lera, T.; Garcia-Diaz, M.; Gonzalez, M.A.; Kirchhoff, T.; Martinez, A.C.; Bernad, A.; Blanco, L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000, 19, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; Ling, H.; Woodgate, R.; Yang, W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005, 24, 2957–2967. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Baruch-Torres, N.; Iwai, S.; Herrmann, G.K.; Brieba, L.G.; Yin, Y.W. Human Mitochondrial DNA Polymerase Metal Dependent UV Lesion Bypassing Ability. Front. Mol. Biosci. 2022, 9, 808036. [Google Scholar] [CrossRef]
- Garcia-Diaz, M.; Bebenek, K.; Krahn, J.M.; Pedersen, L.C.; Kunkel, T.A. Role of the catalytic metal during polymerization by DNA polymerase lambda. DNA Repair 2007, 6, 1333–1340. [Google Scholar] [CrossRef]
- Frank, E.G.; Woodgate, R. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J. Biol. Chem. 2007, 282, 24689–24696. [Google Scholar] [CrossRef]
- Park, J.W.; Ames, B.N. 7-Methylguanine adducts in DNA are normally present at high levels and increase on aging: Analysis by HPLC with electrochemical detection. Proc. Natl. Acad. Sci. USA 1988, 85, 7467–7470. [Google Scholar] [CrossRef]
- Hu, G.; Tsai, A.L.; Quiocho, F.A. Insertion of an N7-methylguanine mRNA cap between two coplanar aromatic residues of a cap-binding protein is fast and selective for a positively charged cap. J. Biol. Chem. 2003, 278, 51515–51520. [Google Scholar] [CrossRef]
- Haschemeyer, A.E.; Rich, A. Nucleoside conformations: An analysis of steric barriers to rotation about the glycosidic bond. J. Mol. Biol. 1967, 27, 369–384. [Google Scholar] [CrossRef]
- Wilson, H.R.; Rahman, A. Nucleoside conformation and non-bonded interactions. J. Mol. Biol. 1971, 56, 129–142. [Google Scholar] [CrossRef]
- Son, T.D.; Guschlbauer, W.; Gueron, M. Flexibility and conformations of guanosine monophosphates by the Overhauser effect. J. Am. Chem. Soc. 1972, 94, 7903–7911. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.; Vojtechovsky, J.; Clowney, L.; Brunger, A.T.; Berman, H.M. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Kou, Y.; Ouzon-Shubeita, H.; Lee, S. Transition-state destabilization reveals how human DNA polymerase beta proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res. 2014, 42, 8755–8766. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.J.; Suo, Z. Time-Dependent Extension from an 8-Oxoguanine Lesion by Human DNA Polymerase Beta. J. Am. Chem. Soc. 2017, 139, 9684–9690. [Google Scholar] [CrossRef]
- Kimsey, I.J.; Szymanski, E.S.; Zahurancik, W.J.; Shakya, A.; Xue, Y.; Chu, C.C.; Sathyamoorthy, B.; Suo, Z.; Al-Hashimi, H.M. Dynamic basis for dG*dT misincorporation via tautomerization and ionization. Nature 2018, 554, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Lawley, P.D.; Brookes, P. Acidic dissociation of 7:9-dialkylguanines and its possible relation to mutagenic properties of alkylating agents. Nature 1961, 192, 1081–1082. [Google Scholar] [CrossRef]
- Kou, Y.; Koag, M.C.; Lee, S. N7 methylation alters hydrogen-bonding patterns of guanine in duplex DNA. J. Am. Chem. Soc. 2015, 137, 14067–14070. [Google Scholar] [CrossRef]
- Yasui, M.; Suzuki, N.; Miller, H.; Matsuda, T.; Matsui, S.; Shibutani, S. Translesion synthesis past 2′-deoxyxanthosine, a nitric oxide-derived DNA adduct, by mammalian DNA polymerases. J. Mol. Biol. 2004, 344, 665–674. [Google Scholar] [CrossRef]
- Yasui, M.; Suenaga, E.; Koyama, N.; Masutani, C.; Hanaoka, F.; Gruz, P.; Shibutani, S.; Nohmi, T.; Hayashi, M.; Honma, M. Miscoding properties of 2′-deoxyinosine, a nitric oxide-derived DNA Adduct, during translesion synthesis catalyzed by human DNA polymerases. J. Mol. Biol. 2008, 377, 1015–1023. [Google Scholar] [CrossRef]







| DNA Lesion. | Mutagenic Property | References |
|---|---|---|
| Cyclobutane pyrimidine dimer (CPD) | Error-free insertion of dATP across TT | [33] |
| Cisplatin intrastrand crosslink GpG | Error-free insertion of dCTP | [38] |
| T:G Mismatch | Error-prone insertion of dTTP | [51] |
| 8-Oxoguanine (8-oxoG) | Error-prone insertion with 3.5:1 ratio of dCTP:dATP | [52] |
| 8-Oxoadenine (8-oxoA) | Error-prone insertion with 2:1 ratio of dTTP:dGTP | [35] |
| 8,5′-cyclo-2′-deoxyadenosine (cdA) | Error-free insertion of dTTP across cdA (Mg2+) | [53] |
| Abasic (AP) site | Purine nucleotide insertion across AP next to dT/dC | [37] |
| 5-N-methylformamidopyrimidine dG | Error-free insertion of dCTP across FapydG | [54] |
| Ribonucleotide insertion | dCTP:rCTP ratio is 1:0.005 across dG | [55] |
| N7-methylguanine (N7mG) | Error-free insertion with 14:1 ratio of dCTP:dTTP | [39] |
| N7-benzylguanine (N7BnG) | Error-free insertion with 10:1 ratio of dCTP:dTTP | [40] |
| O6-methylguanine (O6mG) | Error-prone insertion with 1:1 ratio of dTTP:dCTP | [50] |
| Xanthine (XT) | Error-prone insertion with 3:1 ratio of dCTP:dTTP | [41] |
| Hypoxanthine (HX) | Exclusive error-prone insertion of dCTP | [41] |
| N7-nitrogen half-mustard (NHMG) | Error-free insertion with 10:1 ratio of dCTP:dTTP | [14] |
| Template:dNTP | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | f a |
|---|---|---|---|---|
| dA:dTTP (Mg2+) | 5.4 ± 0.7 | 109 ± 13 | 20 | 1 |
| cdA:dTTP (Mg2+) | 570 ± 70 | 8.6 ± 0.5 | 0.015 | 6.7 × 10−4 |
| dA:dTTP (Mn2+) | 0.44 ± 0.04 | 82 ± 5 | 186 | 1 |
| cdA:dTTP (Mn2+) | 0.49 ± 0.07 | 10.1 ± 0.2 | 21 | 0.11 |
| Template:dNTP | Km (μM) | kcat (10−3 s−1) | kcat/Km (10−3 s−1 μM−1) | f a |
|---|---|---|---|---|
| dG:dCTP | 2.7 ± 0.3 | 120.6 ± 6.1 | 46 | 1 |
| dG:dTTP | 159.3 ± 2.7 | 74.8 ± 0.9 | 0.5 | 0.01 |
| N7mG:dCTP | 4.3 ± 0.4 | 56.4 ± 2.7 | 13 | 1 |
| N7mG:dTTP | 52.5 ± 1.7 | 49.3 ± 0.1 | 0.9 | 0.07 |
| N7BnG:dCTP | 10.2 ± 2.4 | 20.6 ± 3.6 | 2.1 | 1 |
| N7BnG:dTTP | 51.7 ± 5.3 | 11.5 ± 0.3 | 0.2 | 0.1 |
| N7BnG:dCTP (Mn2+) | 5.6 ± 0.9 | 38.7 ± 4.4 | 6.9 | 1 |
| N7BnG:dTTP (Mn2+) | 18.6 ± 1.9 | 17.8 ± 2.1 | 1.0 | 0.14 |
| NHMG:dCTP | 113.4 ± 1.4 | 40.9 ± 1.3 | 0.36 | 1 |
| NHMG:dTTP | 146.3 ± 5.1 | 5.5 ± 0.1 | 0.037 | 0.1 |
| Template:dNTP | Km (μM) | kcat (10−3 s−1) | kcat/Km (10−3 s−1 μM−1) | f a |
|---|---|---|---|---|
| dG:dCTP | 1.3 ± 0.2 | 1330 ± 50 | 1000 | 1 |
| dG:dATP | 92 ± 23 | 100 ± 10 | 1.1 | 0.001 |
| oxoG:dCTP | 2.3 ± 0.2 | 1200 ± 30 | 520 | 1 |
| oxoG:dATP | 5.4 ± 0.6 | 780 ± 30 | 150 | 0.28 |
| dA:dTTP | 5.4 ± 0.2 | 90.9 ± 5.8 | 17 | 1 |
| dA:dGTP | 76.3 ± 4.8 | 6.3 ± 0.5 | 0.08 | 0.005 |
| oxoA:dTTP | 3.6 ± 0.3 | 37.3 ± 2.3 | 11 | 1 |
| oxoA:dGTP | 4.9 ± 0.3 | 24.8 ± 1.3 | 5.1 | 0.46 |
| oxoA:dTTP (Q38A) | 71.3 ± 4.4 | 172.6 ± 5.6 | 2.4 | 1 |
| oxoA:dGTP (Q38A) | 113.8 ± 2.8 | 10.5 ± 0.1 | 0.092 | 0.037 |
| Template:dNTP | Km (μM) | kcat (10−3 s−1) | kcat/Km (10−3 s−1 μM−1) | f a |
|---|---|---|---|---|
| dG:dCTP | 2.7 ± 0.3 | 120.6 ± 6.1 | 46 | 1 |
| dG:dTTP | 159.3 ± 2.7 | 74.8 ± 0.9 | 0.5 | 0.01 |
| XT:dCTP | 10.7 ± 0.9 | 123.3 ± 3.6 | 11.5 | 1 |
| XT:dTTP | 20.6 ± 0.9 | 82.2 ± 4.2 | 4.0 | 0.34 |
| dA:dTTP | 5.4 ± 0.2 | 90.9 ± 5.8 | 17 | 1 |
| dA:dCTP | 80.3 ± 3.2 | 15.2 ± 2.5 | 0.19 | 0.011 |
| HX:dTTP | 21.9 ± 1.4 | 11.7 ± 0.2 | 0.54 | 1 |
| HX:dCTP | 4.6 ± 0.4 | 170.5 ± 4.1 | 37.4 | 69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H. Contributing Factors for Mutagenic DNA Lesion Bypass by DNA Polymerase Eta (polη). DNA 2022, 2, 205-220. https://doi.org/10.3390/dna2040015
Jung H. Contributing Factors for Mutagenic DNA Lesion Bypass by DNA Polymerase Eta (polη). DNA. 2022; 2(4):205-220. https://doi.org/10.3390/dna2040015
Chicago/Turabian StyleJung, Hunmin. 2022. "Contributing Factors for Mutagenic DNA Lesion Bypass by DNA Polymerase Eta (polη)" DNA 2, no. 4: 205-220. https://doi.org/10.3390/dna2040015