Origin of the Bunun Indigenous People of Taiwan, a Review of Published Material Using Y-Chromosome and Mitochondrial DNA Gene Systems
Abstract
:1. Introduction
2. Results and Discussion
2.1. MtDNA Genetic Diversity and Structure
2.1.1. MtDNA Polymorphism
2.1.2. Principal Component Analysis
2.2. Non-Recombining Y-Chromosome
2.2.1. Y-SNP
2.2.2. Y-STR Haplotype Networks
2.2.3. Principal Component Analysis from NRY Haplogroups
2.2.4. TreeMix
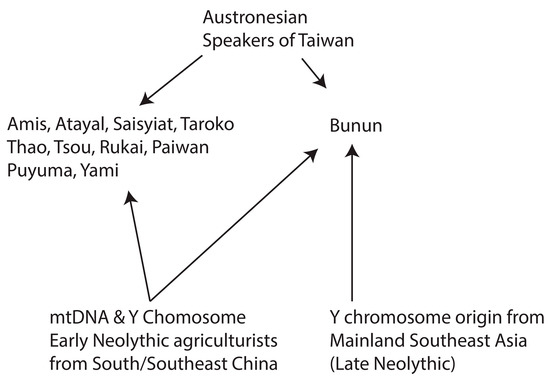
2.3. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MOI. Department of Statistics, Ministry of the Interior, Monthly Bulletin of Interior, Taiwan 2016 Taipei, Taiwan. Available online: https://www.moi.gov.tw (accessed on 17 July 2022).
- Bellwood, P. The Origin and Dispersals and Dispersal of Agricultural Communities in Southest Asia. In Southeast Asia: From Prehistory to History; Glover, I., Bellwood, P., Eds.; Routledge: Abingdon, UK, 2004; pp. 21–40. [Google Scholar]
- Liu, L.; Wang, J.; Levin, M.J.; Sinnott-Amstrong, N.; Zhao, H.; Zhao, Y.; Shao, J.; Di, N.; Zhang, T. The origins of specialized pottery and diverse alcohol fermentation techniques in Early Neolithic China. Proc. Natl. Acad. Sci. USA. 2019, 116, 12767–12774. [Google Scholar] [CrossRef]
- Tsang, C.-H. Five Thousand Years of Taiwan’s Past Brought to Light by Rescue Archaeology. 2015. Available online: http://www.chinesearchaeology.net/en/Special_Events/dierjieshanghailuntan/2015/1222/52517.html (accessed on 7 July 2022).
- Hung, H.; Nguyen, K.D.; Bellwood, P.; Carson, M.T. Coastal Connectivity: Long-Term Trading Networks Across the South China Sea. J. Isl. Coast. Archaeol. 2013, 8, 384–404. [Google Scholar] [CrossRef]
- Liu, Y.C.; Wang, S. Encountering the wider world before the transition to history: Chinese ceramics in proto-historic Taiwan (tenth through sixteenth centuries). In Historical Archaeology of Early Modern Colonialism in Asia-Pacific: The Southwest Pacific and Oceanian Regions; Berrocal, M.C., Tsang, C., Eds.; University Press of Florida: Gainesville, FL, USA, 2017; pp. 270–391. [Google Scholar]
- Ross, M. Proto Austronesian verbal morphology: A reappraisal. In Austronesian Historical Linguistics and Culture History: A Festschrift for Robert Blust; Adelaar, A.P.A., Ed.; Pacific Linguistics: Canberra, Australia, 2009; pp. 295–326. [Google Scholar]
- DGBAS—Directorate General of Budget, Accounting and Statistics, Executive Yuan, R.O.C. National Statistics, (Taiwan). Preliminary Statistical Analysis Report of 2000 Population and Housing Census Archived 12 March 2007 at the Wayback Machine; Excerpted from Table 28: Indigenous Population Distribution in Taiwan-Fukien Area. 2007. Available online: https://eng.dgbas.gov.tw/ (accessed on 7 June 2022).
- Chiang, B. Customary Laws, Kinship and Beyond: A Critical Review of the cultural Anthropological studies of the Austronesian People in Taiwan: In Austronesian Taiwan: Linguistic, History, Ethnology, and Prehistory; Blundel, D., Ed.; The Phoebe Museum of Anthropology: Berkeley, CA, USA, 2002; pp. 201–245. [Google Scholar]
- Huang, Y.-K. The “Great Men” model among the Bunun of Taiwan. In Austronesian Studies Relating to Taiwan; Paul, L.J., Tsang, C., Huang, Y., Ho, D., Tseng, C., Eds.; Academica Sinica: Taipei, Taiwan, 1995; pp. 59–107. [Google Scholar]
- Jacobs, J.B. Taiwan’s Colonial Experiences and the Development of Ethnic Identities: Some Hypotheses; Taiwan in Comparative Perspective, Asia Research Centre, London School of Economics and Political Science: London, UK, 2014; Volume 5, pp. 47–59. ISSN 1752–7732. [Google Scholar]
- Nakamura, T. Household registration of indigenous villages during the Dutch occupation. In Taiwan History during the Dutch Occupation; Wu, T.-M., Wong, K.-I., Shu, S.-Y., Eds.; National Library of Taiwan; Academia Sinica: Taipei, Taiwan, 2002; Volume 11. (In Chinese) [Google Scholar]
- Brown, S.; Savage, P.E.; Ko, A.M.-S.; Stoneking, M.; Ko, Y.-C.; Loo, J.-H.; Trejaut, J.A. Correlations in the population structure of music, genes and language. Proc. Biol. Sci. 2014, 281, 20132072. [Google Scholar] [CrossRef] [PubMed]
- Li, P.J.-K. Bulletin of the Institute of History and Philology. A comparative study of Bunun dialects, Taiwan. Acad. Sin. 1988, 59, 479–508. [Google Scholar]
- De Busser, R.L.J. Towards a Grammar of Takivatan Bunun. Ph.D. Thesis, La Trobe University, Bundoora, Victoria, Australia, 2009. [Google Scholar]
- Umetsu, K.; Yuasa, I.; Suzuki, T.; Sun, C.S.; Pan, I.H.; Ishida, T.; Saitou, N.; Horai, S. Polymorphisms of complement component I and C1R subcomponent of C1 in nine aboriginal Taiwanese populations. Hum. Biol. 1994, 66, 339–348. [Google Scholar]
- Lin, M. Blood groups and transfusion medicine in Taiwan. J. Formos. Med. Assoc. 1997, 96, 933–942. [Google Scholar] [PubMed]
- Chen, W.J.; Chen, C.H.; Huang, J.; Hsu, Y.P.; Seow, S.V.; Chen, C.C.; Cheng, A.T. Genetic polymorphisms of the promoter region of dopamine D2 receptor and dopamine transporter genes and alcoholism among four aboriginal groups and Han Chinese in Taiwan. Psychiatr. Genet. 2002, 11, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-M.; Su, C.-W.; Tsai, L.-C.; Huang, N.-E.; Shih, R.T.-P.; Linacre, A. Analysis of the Alu insertion polymorphism in Taiwanese Han population. Forensic Sci. J. 2002, 1, 27–30. [Google Scholar]
- Chu, C.C.; Lin, M.; Nakajima, F.; Lee, H.L.; Chang, S.L.; Juji, T.; Tokunaga, K. Diversity of HLA among Taiwan’s indigenous tribes and the Ivatans in the Philippines. Tissue Antigens 2001, 58, 9–18. [Google Scholar] [CrossRef]
- Lin, M.; Chu, C.C.; Lee, H.L.; Chang, S.L.; Ohashi, J.; Tokunaga, K.; Akaza, T.; Juji, T. Heterogeneity of Taiwan’s indigenous population: Possible relation to prehistoric Mongoloid dispersals. Tissue Antigens 2000, 55, 1–9. [Google Scholar] [CrossRef]
- Trejaut, J.A.; Muyard, F.; Lai, Y.-H.; Chen, L.-R.; Chen, Z.-S.; Loo, J.-H.; Huang, J.-Y.; Lin, M. Genetic diversity of the Thao people of Taiwan using Y-chromosome, mitochondrial DNA and HLA gene systems. BMC Evol. Biol. 2019, 19, 64. [Google Scholar] [CrossRef]
- Shinoda, K.-I. MtDNA Analysis of Bunun Remains Stored in the National Taiwan University. Anthropol. Sci. 2008, 116, 154–160. [Google Scholar] [CrossRef]
- Tajima, A.; Sun, C.-S.; Pan, I.-H.; Ishida, T.; Saitou, N.; Horai, S. Mitochondrial DNA polymorphisms in nine aboriginal groups of Taiwan: Implications for the population history of aboriginal Taiwanese. Hum. Genet. 2003, 113, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Trejaut, J.A.; Poloni, E.S.; Yen, J.-C.; Lai, Y.-H.; Loo, J.-H.; Lee, C.-L.; He, C.-L.; Lin, M. Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia. BMC Genet. 2014, 15, 77. [Google Scholar] [CrossRef]
- Huang, J.-Y.; Trejaut, J.A.; Lee, C.-L.; Wang, T.-Y.; Loo, J.-H.; Chen, Z.S.; Chen, L.R.; Liu, K.H.; Liu, Y.C.; Hu, C.H.; et al. Mitochondrial DNA Sequencing of Middle Neolithic Human Remains of Ling-Ding Site II: Implication for the Social Structure and the Origin of Northeast Coast Taiwaneses. J. Phylogenetics Evol. Biol. 2018, 6, 2. [Google Scholar] [CrossRef]
- Chen, L.-R.; Trejaut, J.A.; Lai, Y.-H.; Chen, Z.-S.; Huang, J.-Y.; Lin, M.; Loo, J.-H. Mitochondrial DNA Polymorphisms of the Saisiyat Indigenous Group of Taiwan. Search for a Negrito Signature. Edelweiss J. Biomed. Res. Rev. 2019, 12, 12–18. [Google Scholar] [CrossRef]
- Chen, Z.-S.; Trejaut, J.; Loo, J.-H.; Lai, Y.-H.; Huang, J.-Y.; Lin, M. Mitochondrial DNA Diversity of the Nangan Islanders Living in the Mazu Archipelago of the Taiwan Strait. Edelweiss J. Biomed. Res. 2021, 3, 25–27. [Google Scholar]
- Van Oven, M.; Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef]
- Loo, J.-H.; Trejaut, J.A.; Yen, J.-C.; Chen, Z.-S.; Ng, W.-M.; Huang, C.-Y.; Hsu, K.-N.; Hung, K.-H.; Hsiao, Y.; Wei, Y.-H.; et al. Mitochondrial DNA association study of type 2 diabetes with or without ischemic stroke in Taiwan. BMC Res. Notes 2014, 7, 223. [Google Scholar] [CrossRef]
- Trejaut, J.A.; Kivisild, T.; Loo, J.H.; Lee, C.L.; He, C.L.; Hsu, C.J.; Li, Z.Y.; Lin, M. Traces of archaic mitochondrial lineages persist in Austronesian-speaking Formosan populations. PLoS Biol. 2005, 3, e247. [Google Scholar] [CrossRef]
- Li, H.; Wen, B.; Chen, S.J.; Su, B.; Pramoonjago, P.; Liu, Y.; Pan, S.; Qin, Z.; Liu, W.; Cheng, X.; et al. Paternal genetic affinity between Western Austronesians and Daic populations. BMC Evol. Biol. 2008, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.M.; Chen, C.Y.; Fu, Q.; Delfin, F.; Li, M.; Chiu, H.L.; Stoneking, M.; Ko, Y.C. Early Austronesians: Into and out of Taiwan. Am. J. Hum. Genet. 2014, 94, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Melton, T.; Clifford, S.; Martinson, J.; Batzer, M.; Stoneking, M. Genetic evidence for the proto-Austronesian homeland in Asia: mtDNA and nuclear DNA variation in Taiwanese aboriginal tribes. Am. J. Hum. Genet. 1998, 63, 1807–1823. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.-C.; Chen, M.-Y.; Chao, C.-H.; Pu, C.-E. Study on the genetic polymorphisms of Y chromosomal DNA short tandem repeat loci applied to analyzing the relative affinities among ethnic groups in Taiwan. Forensic Sci. Int. Genet. Supplement. Series 2013, 4, e69–e70. [Google Scholar] [CrossRef]
- Xue, Y.; Zerjal, T.; Bao, W.; Zhu, S.; Shu, Q.; Xu, J.; Du, R.; Fu, S.; Li, P.; Hurles, M.E.; et al. Male demography in East Asia: A north-south contrast in human population expansion times. Genetics. 2006, 172, 2431–2439. [Google Scholar] [CrossRef]
- Luo, X.Q.; Du, P.X.; Wang, L.X.; Zhou, B.Y.; Li, Y.C.; Zheng, H.X.; Wei, L.H.; Liu, J.J.; Sun, C.; Meng, H.L.; et al. Uniparental Genetic Analyses Reveal the Major Origin of Fujian Tanka from Ancient Indigenous Daic Populations. Hum. Biol. 2020, 91, 257–277. [Google Scholar] [CrossRef]
- Loo, J.H.; Trejaut, J.A.; Yen, J.C.; Chen, Z.S.; Lee, C.L.; Lin, M. Genetic affinities between the Yami tribe people of Orchid Island and the Philippine Islanders of the Batanes archipelago. BMC Genet. 2011, 12, 21. [Google Scholar] [CrossRef]
- Peng, M.S.; He, J.D.; Liu, H.X.; Zhang, Y.P. Tracing the legacy of the early Hainan Islanders—A perspective from mitochondrial DNA. BMC Evol. Biol. 2011, 11, 46. [Google Scholar] [CrossRef]
- Kong, Q.P.; Yao, Y.G.; Liu, M.; Shen, S.P.; Chen, C.; Zhu, C.L.; Palanichamy, M.G.; Zhang, Y.P. Mitochondrial DNA sequence polymorphisms of five ethnic populations from northern China. Hum. Genet. 2003, 113, 391–405. [Google Scholar] [CrossRef]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Research. 2019, 48, D941–D947. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.D.; Sun, Y.B.; Li, E.M.; Xu, L.Y.; Zhang, Y.P.; Yao, Y.G.; Kong, Q.P. Deciphering the signature of selective constraints on cancerous mitochondrial genome. Mol. Biol. Evol. 2012, 29, 1255–1261. [Google Scholar] [CrossRef]
- Kong, Q.-P.; Yao, Y.-G.; Sun, C.; Bandelt, H.-J.; Zhu, C.-L.; Zhang, Y.-P. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 2003, 73, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Delfin, F.; Min-Shan Ko, A.; Li, M.; Gunnarsdóttir, E.D.; Tabbada, K.A.; Salvador, J.M.; Calacal, G.C.; Sagum, M.S.; Datar, F.A.; Padilla, S.G.; et al. Complete mtDNA genomes of Filipino ethnolinguistic groups: A melting pot of recent and ancient lineages in the Asia-Pacific region. Eur. J. Hum. Genet. 2014, 22, 228–237. [Google Scholar] [CrossRef]
- Duong, N.T.; Macholdt, E.; Ton, N.D.; Arias, L.; Schröder, R.; Van Phong, N.; Thi Bich Thuy, V.; Hai Ha, N.; Thi Thu Hue, H.; Thi Xuan, N.; et al. Complete human mtDNA genome sequences from Vietnam and the phylogeography of Mainland Southeast Asia. Sci. Rep. 2018, 8, 11651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutanan, W.; Kampuansai, J.; Brunelli, A.; Ghirotto, S.; Pittayaporn, P.; Ruangchai, S.; Schröder, R.; Macholdt, E.; Srikummool, M.; Kangwanpong, D.; et al. New insights from Thailand into the maternal genetic history of Mainland Southeast Asia. Eur. J. Hum. Genet. 2018, 26, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Soares, P.; Rito, T.; Trejaut, J.; Mormina, M.; Hill, C.; Tinkler-Hundal, E.; Braid, M.; Clarke, D.J.; Loo, J.H.; Thomson, N.; et al. Ancient voyaging and Polynesian origins. Am. J. Hum. Genet. 2011, 88, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Soares, P.; Mormina, M.; Macaulay, V.; Meehan, W.; Blackburn, J.; Clarke, D.; Raja, J.M.; Ismail, P.; Bulbeck, D.; et al. Phylogeography and Ethnogenesis of Aboriginal Southeast Asians. Mol. Biol. Evol. 2006, 23, 2480–2491. [Google Scholar] [CrossRef]
- Gunnarsdóttir, E.D.; Nandineni, M.R.; Li, M.; Myles, S.; Gil, D.; Pakendorf, B.; Stoneking, M. Larger mitochondrial DNA than Y-chromosome differences between matrilocal and patrilocal groups from Sumatra. Nat. Commun. 2011, 2, 228. [Google Scholar] [CrossRef]
- Jinam, T.A.; Hong, L.C.; Phipps, M.E.; Stoneking, M.; Ameen, M.; Edo, J.; HUGO Pan-Asian SNP Consortium; Saitou, N. Evolutionary history of continental southeast Asians: "early train" hypothesis based on genetic analysis of mitochondrial and autosomal DNA data. Mol. Biol. Evol. 2012, 29, 3513–3527. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Duggan, A.T.; Evans, B.; Friedlaender, F.R.; Friedlaender, J.S.; Koki, G.; Merriwether, D.A.; Kayser, M.; Stoneking, M. Maternal history of Oceania from complete mtDNA genomes: Contrasting ancient diversity with recent homogenization due to the Austronesian expansion. Am. J. Hum. Genet. 2014, 94, 721–733. [Google Scholar] [CrossRef]
- Soares, P.A.; Trejaut, J.A.; Rito, T.; Cavadas, B.; Hill, C.; Eng, K.K.; Mormina, M.; Brandão, A.; Fraser, R.M.; Wang, T.Y.; et al. Resolving the ancestry of Austronesian-speaking populations. Hum. Genet. 2016, 135, 309–326. [Google Scholar] [CrossRef]
- Zhang, X.; Qi, X.; Yang, Z.; Serey, B.; Sovannary, T.; Bunnath, L.; Aun, H.S.; Samnom, H.; Zhang, H.; Lin, Q.; et al. Analysis of mitochondrial genome diversity identifies new and ancient maternal lineages in Cambodian aborigines. Nat. Commun. 2013, 4, 2599. [Google Scholar] [CrossRef] [PubMed]
- Brandão, A.; Eng, K.K.; Rito, T.; Cavadas, B.; Bulbeck, D.; Gandini, F.; Pala, M.; Mormina, M.; Hudson, B.; White, J.; et al. Quantifying the legacy of the Chinese Neolithic on the maternal genetic heritage of Taiwan and Island Southeast Asia. Hum. Genet. 2016, 135, 363–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delfin, F.; Salvador, J.M.; Calacal, G.C.; Perdigon, H.B.; Tabbada, K.A.; Villamor, L.P.; Halos, S.C.; Gunnarsdóttir, E.; Myles, S.; Hughes, D.A.; et al. The Y-chromosome landscape of the Philippines: Extensive heterogeneity and varying genetic affinities of Negrito and non-Negrito groups. Eur. J. Hum. Genet. 2010, 19, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Heyer, E.; Georges, M.; Pachner, M.; Endicott, P. Genetic diversity of four Filipino negrito populations from Luzon: Comparison of male and female effective population sizes and differential integration of immigrants into Aeta and Agta communities. Hum. Biol. 2013, 85, 189–208. [Google Scholar] [CrossRef]
- Su, B.; Jin, L.; Underhill, P.; Martinson, J.; Saha, N.; McGarvey, S.T.; Shriver, M.D.; Chu, J.; Oefner, P.; Chakraborty, R.; et al. Polynesian origins: Insights from the Y chromosome. Proc. Natl. Acad. Sci. USA 2000, 97, 8225–8228. [Google Scholar] [CrossRef]
- Zhong, H.; Shi, H.; Qi, X.B.; Duan, Z.Y.; Tan, P.P.; Jin, L.; Su, B.; Ma, R.Z. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route. Mol. Biol. Evol. 2011, 28, 717–727. [Google Scholar] [CrossRef]
- Kayser, M.; Choi, Y.; van Oven, M.; Mona, S.; Brauer, S.; Trent, R.J.; Suarkia, D.; Schiefenhövel, W.; Stoneking, M. The impact of the Austronesian expansion: Evidence from mtDNA and Y chromosome diversity in the Admiralty Islands of Melanesia. Mol. Biol. Evol. 2008, 25, 1362–1374. [Google Scholar] [CrossRef]
- Urasin, V. YTREE V8.04. Available online: https://www.yfull.com/arch-8.04/tree/ (accessed on 7 July 2022).
- Waas, M.; Yacobi, D.; Kull, L.; Urasin, V.; Magoon, G.; Penninx, W.; Brown, A.; Nogueiro, I. Haplogroup J-Z640-Genetic Insight into the Levantine Bronze Age. J. Phylogenetics Evol. Biol. 2019, 7, 209. [Google Scholar]
- Razafindrazaka, H. Le Peuplement Humain de Madagascar: Anthropologie Génétique de Trois Groupes Traditionnels; Université Toulouse III-Paul Sabatier: Toulouse, France, 2010. (In French) [Google Scholar]
- Razafindrazaka, H.; Ricaut, F.X.; Cox, M.P.; Mormina, M.; Dugoujon, J.M.; Randriamarolaza, L.P.; Guitard, E.; Tonasso, L.; Ludes, B.; Crubézy, E. Complete mitochondrial DNA sequences provide new insights into the Polynesian motif and the peopling of Madagascar. Eur. J. Hum. Genet. 2010, 18, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Tofanelli, S.; Ferri, G.; Bulayeva, K.; Caciagli, L.; Onofri, V.; Taglioli, L.; Bulayev, O.; Boschi, I.; Alù, M.; Berti, A.; et al. J1-M267 Y lineage marks climate-driven pre-historical human displacements. Eur. J. Hum. Genet. 2009, 17, 1520–1524. [Google Scholar] [CrossRef] [PubMed]
- van Oven, M.; Hammerle, J.M.; van Schoor, M.; Kushnick, G.; Pennekamp, P.; Zega, I.; Lao, O.; Brown, L.; Kennerknecht, I.; Kayser, M. Unexpected island effects at an extreme: Reduced Y chromosome and mitochondrial DNA diversity in Nias. Mol. Biol. Evol. 2011, 28, 1349–1361. [Google Scholar] [CrossRef] [PubMed]
- He, J.D.; Peng, M.S.; Quang, H.H.; Dang, K.P.; Trieu, A.V.; Wu, S.F.; Jin, J.Q.; Murphy, R.W.; Yao, Y.G.; Zhang, Y.P. Patrilineal perspective on the Austronesian diffusion in Mainland Southeast Asia. PLoS ONE 2012, 7, e36437. [Google Scholar] [CrossRef]
- Cai, X.; Qin, Z.; Wen, B.; Xu, S.; Wang, Y.; Lu, Y.; Wei, L.; Wang, C.; Li, S.; Huang, X.; et al. Human migration through bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum revealed by Y chromosomes. PLoS ONE 2011, 6, e24282. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Kampuansai, J.; Qi, X.; Yan, S.; Yang, Z.; Serey, B.; Sovannary, T.; Bunnath, L.; Aun, H.S.; Samnom, H.; et al. An Updated Phylogeny of the Human Y-Chromosome Lineage O2a-M95 with Novel SNPs. PLoS ONE 2014, 9, e101020. [Google Scholar] [CrossRef]
- Dalton, R. Neanderthal DNA yields to genome foray. Nature 2006, 441, 260–261. [Google Scholar] [CrossRef]
- Brunelli, A.; Kampuansai, J.; Seielstad, M.; Lomthaisong, K.; Kangwanpong, D.; Ghirotto, S.; Kutanan, W. Y chromosomal evidence on the origin of northern Thai people. PLoS ONE 2017, 12, e0181935. [Google Scholar]
- Simonson, T.S.; Xing, J.; Barrett, R.; Jerah, E.; Loa, P.; Zhang, Y.; Watkins, W.S.; Witherspoon, D.J.; Huff, C.D.; Woodward, S.; et al. Ancestry of the Iban is predominantly Southeast Asian: Genetic evidence from autosomal, mitochondrial, and Y chromosomes. PLoS ONE 2011, 6, e16338. [Google Scholar] [CrossRef]
- Blust, R. Subgrouping, circularity and extinction: Some issues in Austronesian comparative linguistics. Symp. Ser. Inst. Linguist. Acad. Sinica 1999, 1, 31–94. [Google Scholar]
- Msaidie, S.; Ducourneau, A.; Boetsch, G.; Longepied, G.; Papa, K.; Allibert, C.; Yahaya, A.A.; Chiaroni, J.; Mitchell, M.J. Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean. Eur. J. Hum. Genet. 2011, 19, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Tofanelli, S.; Bertoncini, S.; Castri, L.; Luiselli, D.; Calafell, F.; Donati, G.; Paoli, G. On the origins and admixture of Malagasy: New evidence from high-resolution analyses of paternal and maternal lineages. Mol. Biol. Evol. 2009, 26, 2109–2124. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Li, H. Inferring human history in East Asia from Y chromosomes. Investig. Genet. 2013, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- ISSOG. International Society of Genetic Genealogy. Y-DNA Haplogroup Tree 2019, Version 15.46. Available online: https://www.isogg.org/tree/ (accessed on 12 March 2020).
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef]
- Soares, P.; Trejaut, J.A.; Loo, J.H.; Hill, C.; Mormina, M.; Lee, C.L.; Chen, Y.M.; Hudjashov, G.; Forster, P.; Macaulay, V.; et al. Climate change and postglacial human dispersals in southeast Asia. Mol. Biol. Evol. 2008, 25, 1209–1218. [Google Scholar] [CrossRef]
- Tabbada, K.A.; Trejaut, J.; Loo, J.H.; Chen, Y.M.; Lin, M.; Mirazon-Lahr, M.; Kisivild, T.; De Ungria, M.C.A. Philippine mitochondrial DNA diversity: A populated viaduct between Taiwan and Indonesia? Mol. Biol. Evol. 2010, 27, 21–31. [Google Scholar] [CrossRef]
- Kutanan, W.; Kampuansai, J.; Srikummool, M.; Brunelli, A.; Ghirotto, S.; Arias, L.; Macholdt, E.; Hübner, A.; Schröder, R.; Stoneking, M. Contrasting Paternal and Maternal Genetic Histories of Thai and Lao Populations. Mol. Biol. Evol. 2019, 36, 1490–1506. [Google Scholar] [CrossRef]
- Gan, R.J.; Pan, S.L.; Mustavich, L.F.; Qin, Z.D.; Cai, X.Y.; Qian, J.; Liu, C.W.; Peng, J.H.; Li, S.L.; Xu, J.S.; et al. Pinghua population as an exception of Han Chinese’s coherent genetic structure. J. Hum. Genet. 2008, 53, 303–313. [Google Scholar] [CrossRef]
- Li, H. Genetic Structure of Austro-Tai Populations. Ph.D. Dissertation, Fudan University, Shanghai, China, 2005. [Google Scholar]
- Kutanan, W.; Kampuansai, J.; Srikummool, M.; Kangwanpong, D.; Ghirotto, S.; Brunelli, A.; Stoneking, M. Complete mitochondrial genomes of Thai and Lao populations indicate an ancient origin of Austroasiatic groups and demic diffusion in the spread of Tai-Kadai languages. Hum Genet. 2017, 136, 85–98. [Google Scholar] [CrossRef]
- Li, D.; Li, H.; Ou, C.; Lu, Y.; Sun, Y.; Yang, B.; Qin, Z.; Zhou, Z.; Li, S.; Jin, L. Paternal genetic structure of Hainan aborigines isolated at the entrance to East Asia. PLoS ONE 2008, 3, e2168. [Google Scholar] [CrossRef]
- Bekaert, B.; Zainuddin, Z.; Hadi, S.; Goodwin, W. A comparison of mtDNA and Y chromosome diversity in Malay populations. Int. Congr. Ser. 2006, 1288, 252–255. [Google Scholar] [CrossRef]
- Chen, Y.; Chu, J.Y.; Yu, J.K.; Yu, L.; Sun, H.; Lin, K.Q.; Tao, Y.F.; Shi, L.; Huang, X.Q.; Shi, T.L.; et al. Polymorphism of DYS287 on Y chromosome in 28 ethnic populations of China. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2006, 28, 196–201. (In Chinese) [Google Scholar] [PubMed]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstätter, A.; Forer, L.; Specht, G.; Bandelt, H.; Kronenberg, F.; Salas, A.; Schönherr, S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucl. Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. Available online: http://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed on 17 July 2022).
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Zhivotovsky, L.A.; Underhill, P.A.; Cinnioğlu, C.; Kayser, M.; Morar, B.; Kivisild, T.; Scozzari, R.; Cruciani, F.; Destro-Bisol, G.; Spedini, G.; et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004, 74, 50–61. [Google Scholar] [CrossRef] [Green Version]






| Country | Population | Size | Languages | Groups | References | ||
|---|---|---|---|---|---|---|---|
| MtDNA | NRY | MtDNA | NRY | ||||
| Taiwan | Bunun (Northern) | 89 | 56 | Austronesian | Northern AN_Tw | [31] | [22,25,32] |
| Taiwan | Bunun (Central) | 50 | n/a | Austronesian | Central AN_Tw | [33] | n/a |
| Taiwan | Bunun (Miscellaneous) | 96 | n/a | Austronesian | Miscellaneous | [24,34] | [35] (STRs only) |
| Taiwan | Hakka | 45 | 34 | Sinitic | TwH/Urban Taiwanese | [33] | [25] |
| Taiwan | Minnan | 50 | 60 | Sinitic | TwH/Urban Taiwanese | [33] | [25,36] |
| China | Fujian and Matsu | 198 | 55 | Sinitic | East Coast China | [28,37] | [25] |
| Taiwan | Saisiyat | 87 | 24 | Austronesian | Northern AN_Tw | [31,33] | [25] |
| Taiwan | Atayal | 157 | 52 | Austronesian | Northern AN_Tw | [31,33] | [25] |
| Taiwan | Amis | 148 | 39 | Austronesian | East Coast Taiwan | [31,33] | [25] |
| Taiwan | Taroko | 54 | 20 | Austronesian | Northern AN_Tw | [22] * | [25] |
| Taiwan | Thao | 30 | 16 | Austronesian | Central AN_Tw | [22,31,33] | [22,25] |
| Taiwan | Tsou | 108 | 41 | Austronesian | Central AN_Tw | [31,33] | [25] |
| Taiwan | Paiwan | 105 | 25 | Austronesian | Southern AN_Tw | [31,33] | [25] |
| Taiwan | Puyuma | 91 | 23 | Austronesian | Southern AN_Tw | [31,33] | [25] |
| Taiwan | Rukai | 99 | 29 | Austronesian | Southern AN_Tw | [31,33] | [25] |
| Taiwan | Yami | 123 | 30 | Austronesian | Southern AN_Tw | [31,38] | [25,38] |
| Northeast China | Beijing, Henan, and Liaoning | 257 | na | Sinitic | NE_China (Han) | [39,40,41,42,43] | [36] |
| South China | South China | 65 | na | Sinitic | Sth_China (Han) | [41,43] | [32,36] |
| Philippines | Indigenous groups | 260 | 121 | Malayo Polynesian | ISEA | [44] | [25] |
| Vietnam | Indigenous groups | 603 | 24 | Austro Asiatic | MSEA | [45] | [25] |
| Thailand | Indigenous groups | 560 | 75 | Kra-Dai | MSEA | [46] | [25] |
| Indonesia (East) | East_Indonesia | 72 | na | Malayo Polynesian | ISEA | [47,48,49] | [25,32] |
| Indonesia (West) | West_Indonesia | 326 | 68 | Malayo Polynesian | ISEA | [47,48,49] | [25] |
| Malaysia | Indigenous groups | 86 | 8 | Austronesian | MSEA | [50] | [25] |
| Total | 4423 | 800 | |||||
| Populations | China | Taiwan | Island Southeast Asia | Mainland Southeast Asia | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Han | Urban Taiwanese | Taiwan Indigenous People (AN_Tw) | Philippines | Indonesia | Total Indonesia (East, West and 36 undefined) | Thailand Total | Vietnam (Hanoy) | |||||||||||||
| Fujian | Hakka | Minnan | Atayal | Taroko | Saisiyat | Thao | Northern Bunun | Tsou | Yami | Amis | Rukai | Paiwan | Puyuma | West Indonesia (Java+Borneo+Sumatra) | East Indonesia(Maluku Ambon) | |||||
| O1a2M50 | 2.94 | 1.92 | 4.17 | 6.25 | 60.71 | 4.88 | 17.95 | 24.14 | 28.00 | 21.74 | 11.64 | 5.73 | 11.11 | 5.28 | ||||||
| O1b1a1a1a1a1-M88 (O2a1aM88) | 2.94 | 1.67 | 37.50 | 3.33 | 3.42 | 3.64 | 3.25 | 19.61 | 25.00 | |||||||||||
| P*M45 | 1.79 | 2.56 | ||||||||||||||||||
| others | 100 | 94.12 | 98.33 | 98.08 | 100 | 95.83 | 93.75 | 0.00 | 95.12 | 96.67 | 79.49 | 75.86 | 72.00 | 78.26 | 84.94 | 90.63 | 88.89 | 91.47 | 80.39 | 75.00 |
| Y-SNP Sample size | 55 | 34 | 60 | 52 | 20 | 24 | 16 | 56 | 41 | 30 | 39 | 29 | 25 | 23 | 146 | 192 | 18 | 246 | 102 | 24 |
| Haplogroup Diversity | 0.86 | 0.92 | 0.90 | 0.18 | 0.10 | 0.24 | 0.24 | 0.50 | 0.19 | 0.65 | 0.69 | 0.48 | 0.73 | 0.72 | 0.90 | 0.86 | 0.88 | 0.87 | 0.89 | 0.91 |
| ±SD | 0.035 | 0.027 | 0.024 | 0.202 | 0.196 | 0.195 | 0.219 | 0.106 | 0.207 | 0.097 | 0.069 | 0.187 | 0.064 | 0.104 | 0.018 | 0.034 | 0.030 | 0.030 | 0.023 | 0.032 |
| Total Number of Haplogroups | 13 | 18 | 16 | 3 | 2 | 4 | 3 | 3 | 3 | 5 | 5 | 3 | 5 | 5 | 26 | 24 | 7 | 27 | 21 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trejaut, J.A. Origin of the Bunun Indigenous People of Taiwan, a Review of Published Material Using Y-Chromosome and Mitochondrial DNA Gene Systems. DNA 2022, 2, 185-201. https://doi.org/10.3390/dna2030013
Trejaut JA. Origin of the Bunun Indigenous People of Taiwan, a Review of Published Material Using Y-Chromosome and Mitochondrial DNA Gene Systems. DNA. 2022; 2(3):185-201. https://doi.org/10.3390/dna2030013
Chicago/Turabian StyleTrejaut, Jean A. 2022. "Origin of the Bunun Indigenous People of Taiwan, a Review of Published Material Using Y-Chromosome and Mitochondrial DNA Gene Systems" DNA 2, no. 3: 185-201. https://doi.org/10.3390/dna2030013