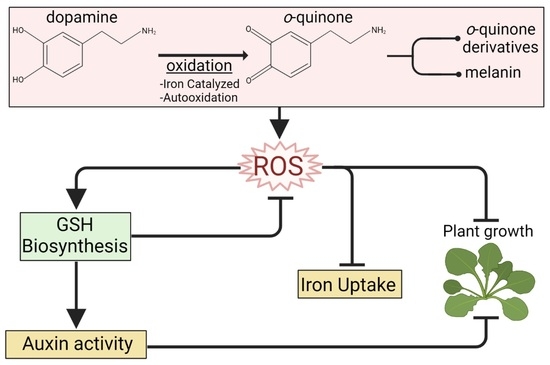
Dopamine Inhibits Arabidopsis Growth through Increased Oxidative Stress and Auxin Activity
Abstract
:1. Introduction
2. Results
2.1. DA Inhibits Arabidopsis Growth and Causes Broad Transcriptomic Changes
2.2. DA Does Not Increase IAA Accumulation
2.3. DA Increases Auxin Action
2.4. DA-Enhanced Auxin Activity Requires the Auxin Transporter AUX1
2.5. DA-Induced Oxidative Stress Elevates the Antioxidant Glutathione
2.6. DA-Enhanced Auxin Activity Depends on Glutathione Synthesis
2.7. DA-Induced Oxidative Stress Suppresses Iron Accumulation
3. Discussion
4. Materials and Methods
4.1. Growth Conditions and Materials
4.2. GUS and Ferric Chelate Reductase (FCR) Assays
4.3. Fluorescent Microscopy
4.4. RNAseq Analysis
4.5. Immunoblotting Analysis
4.6. Auxin Metabolite Profiling
4.7. Iron Determination
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, Q.; Gao, T.; Liu, W.; Liu, Y.; Zhao, Y.; Liu, Y.; Li, W.; Ding, K.; Ma, F.; Li, C. Functions of dopamine in plants: A review. Plant Signal. Behav. 2020, 15, 1827782. [Google Scholar] [CrossRef]
- Berridge, K.C.; Robinson, T.E. What is the role of dopamine in reward: Hedonic impact, reward learning, or incentive salience? Brain Res. Rev. 1998, 28, 309–369. [Google Scholar] [CrossRef]
- Arias-Carrión, O.; Stamelou, M.; Murillo-Rodríguez, E.; Menéndez-González, M.; Pöppel, E. Dopaminergic reward system: A short integrative review. Int. Arch. Med. 2010, 3, 24. [Google Scholar] [CrossRef] [Green Version]
- Kulma, A.; Szopa, J. Catecholamines are active compounds in plants. Plant Sci. 2007, 172, 433–440. [Google Scholar] [CrossRef]
- Zacares, L.; Lopez-Gresa, M.P.; Fayos, J.; Primo, J.; Belles, J.M.; Conejero, V. Induction of p-coumaroyldopamine and feruloyldopamine, two novel metabolites, in tomato by the bacterial pathogen Pseudomonas syringae. Mol. Plant Microbe Interact. 2007, 20, 1439–1448. [Google Scholar] [CrossRef] [Green Version]
- Guidotti, B.B.; Gomes, B.R.; Siqueira-Soares, R.d.C.; Soares, A.R.; Ferrarese-Filho, O. The effects of dopamine on root growth and enzyme activity in soybean seedlings. Plant Signal. Behav. 2013, 8, e25477. [Google Scholar] [CrossRef] [Green Version]
- Soares, A.R.; Marchiosi, R.; de Cássia Siqueira-Soares, R.; Barbosa de Lima, R.; Dantas dos Santos, W.; Ferrarese-Filho, O. The role of L-DOPA in plants. Plant Signal. Behav. 2014, 9, e28275. [Google Scholar] [CrossRef] [Green Version]
- Ahammed, G.J.; Li, X. Dopamine-induced abiotic stress tolerance in horticultural plants. Sci. Hortic. 2023, 307, 111506. [Google Scholar] [CrossRef]
- Protacio, C.M.; Dai, Y.R.; Lewis, E.F.; Flores, H.E. Growth stimulation by catecholamines in plant tissue/organ cultures. Plant Physiol. 1992, 98, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Tsafouros, A.; Roussos, P.A. Dopamine, Chlorogenic Acid, and Quinones as Possible Cofactors of Increasing Adventitious Rooting Potential of In Vitro Krymsk 5 Cherry Rootstock Explants. Agronomy 2022, 12, 1154. [Google Scholar] [CrossRef]
- Swiedrych, A.; Lorenc-Kukula, K.; Skirycz, A.; Szopa, J. The catecholamine biosynthesis route in potato is affected by stress. Plant Physiol. Biochem. 2004, 42, 593–600. [Google Scholar] [CrossRef]
- Kamisaka, S. Catecholamine stimulation of the gibberelin action that induces lettuce hypocotyl elongation. Plant Cell Physiol. 1979, 20, 1199–1207. [Google Scholar] [CrossRef]
- Hayashi, K.-I.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef]
- Meiser, J.; Weindl, D.; Hiller, K. Complexity of dopamine metabolism. Cell Commun. Signal. 2013, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Jodko-Piórecka, K.; Litwinienko, G. Antioxidant activity of dopamine and L-DOPA in lipid micelles and their cooperation with an analogue of α-tocopherol. Free Radic. Biol. Med. 2015, 83, 1–11. [Google Scholar] [CrossRef]
- Spencer, J.P.; Jenner, A.; Butler, J.; Aruoma, O.I.; Dexter, D.T.; Jenner, P.; Halliwell, B. Evaluation of the pro-oxidant and antioxidant actions of L-DOPA and dopamine in vitro: Implications for Parkinson’s disease. Free Radic. Res. 1996, 24, 95–105. [Google Scholar] [CrossRef]
- Bjorklund, G.; Peana, M.; Maes, M.; Dadar, M.; Severin, B. The glutathione system in Parkinson’s disease and its progression. Neurosci. Biobehav. Rev. 2021, 120, 470–478. [Google Scholar] [CrossRef]
- Hare, D.J.; Double, K.L. Iron and dopamine: A toxic couple. Brain 2016, 139, 1026–1035. [Google Scholar] [CrossRef] [Green Version]
- Sulzer, D.; Bogulavsky, J.; Larsen, K.E.; Behr, G.; Karatekin, E.; Kleinman, M.H.; Turro, N.; Krantz, D.; Edwards, R.H.; Greene, L.A.; et al. Neuromelanin biosynthesis is driven by excess cytosolic catecholamines not accumulated by synaptic vesicles. Proc. Natl. Acad. Sci. USA 2000, 97, 11869–11874. [Google Scholar] [CrossRef] [Green Version]
- Gomes, B.R.; Siqueira-Soares, R.d.C.; Dos Santos, W.D.; Marchiosi, R.; Soares, A.R.; Ferrarese-Filho, O. The effects of dopamine on antioxidant enzymes activities and reactive oxygen species levels in soybean roots. Plant Signal. Behav. 2014, 9, e977704. [Google Scholar] [CrossRef] [Green Version]
- Gräfe, K.; Schmitt, L. The ABC transporter G subfamily in Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.J.; Schenk, P.M.; Kazan, K.; Penninckx, I.A.M.A.; Anderson, J.P.; Maclean, D.J.; Cammue, B.P.A.; Ebert, P.R.; Manners, J.M. Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 2003, 133, 1272–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.; Hwang, J.-U.; Lee, M.; Kim, Y.-Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Lee, K.; Lee, J.; Noh, E.W.; Lee, Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 2005, 138, 827–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivitz, A.B.; Hermand, V.; Curie, C.; Vert, G. Arabidopsis bHLH100 and bHLH101 Control Iron Homeostasis via a FIT-Independent Pathway. PLoS ONE 2012, 7, e44843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golisz, A.; Sugano, M.; Hiradate, S.; Fujii, Y. Microarray analysis of Arabidopsis plants in response to allelochemical L-DOPA. Planta 2011, 233, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Luo, R.; Li, J.; Miao, R.; An, H.; Yan, X.; Pang, Q. Arabidopsis glutathione-S-transferases GSTF11 and GSTU20 function in aliphatic glucosinolate biosynthesis. Front. Plant Sci. 2021, 12, 816233. [Google Scholar] [CrossRef]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of Indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef] [Green Version]
- Kumari, V.; Kumar, V.; Bhalla, T.C. Functional interpretation and structural insights of Arabidopsis lyrata cytochrome P450 CYP71A13 involved in auxin synthesis. Bioinformation 2015, 11, 330–335. [Google Scholar] [CrossRef] [Green Version]
- Kissen, R.; Øverby, A.; Winge, P.; Bones, A.M. Allyl-isothiocyanate treatment induces a complex transcriptional reprogramming including heat stress, oxidative stress and plant defence responses in Arabidopsis thaliana. BMC Genom. 2016, 17, 740. [Google Scholar] [CrossRef] [Green Version]
- Goda, H.; Sawa, S.; Asami, T.; Fujioka, S.; Shimada, Y.; Yoshida, S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004, 134, 1555–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravet, K.; Touraine, B.; Boucherez, J.; Briat, J.-F.; Gaymard, F.; Cellier, F. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 2009, 57, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Luhua, S.; Hegie, A.; Suzuki, N.; Shulaev, E.; Luo, X.; Cenariu, D.; Ma, V.; Kao, S.; Lim, J.; Gunay, M.B.; et al. Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant. 2013, 148, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, C.; Ginzburg, D.; Zhao, K.; Dwyer, W.; Xue, B.; Xu, A.; Rice, S.; Cole, B.; Paley, S.; Karp, P.; et al. Plant Metabolic Network 15: A resource of genome-wide metabolism databases for 126 plants and algae. J. Integr. Plant Biol. 2021, 63, 1888–1905. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; Chapman, A.; Fellermeier, F.; Choudhary, M.; Scheel, D.; Glawischnig, E. The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis. Plant Physiol. 2014, 165, 841–853. [Google Scholar] [CrossRef] [Green Version]
- Casanova-Saez, R.; Mateo-Bonmati, E.; Ljung, K. Auxin metabolism in plants. Cold Spring Harb. Perspect. Biol. 2021, 13, a039867. [Google Scholar] [CrossRef]
- Zhao, J.; Williams, C.C.; Last, R.L. Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 1998, 10, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Peer, W.A. Auxin homeostasis: The DAO of catabolism. J. Exp. Bot. 2017, 68, 3145–3154. [Google Scholar] [CrossRef] [Green Version]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 2014, 151, 83–96. [Google Scholar] [CrossRef]
- Kurepa, J.; Shull, T.E.; Smalle, J.A. Antagonistic activity of auxin and cytokinin in shoot and root organs. Plant Direct 2019, 3, e00121. [Google Scholar] [CrossRef] [Green Version]
- Leyser, H.M.; Pickett, F.B.; Dharmasiri, S.; Estelle, M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996, 10, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Delbarre, A.; Muller, P.; Imhoff, V.; Guern, J. Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 1996, 198, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.; Feldmann, K.A. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Friml, J.; Marchant, A.; Ljung, K.; Sandberg, G.; Palme, K.; Bennett, M. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001, 15, 2648–2653. [Google Scholar] [CrossRef] [Green Version]
- Carrier, D.J.; Bakar, N.T.; Swarup, R.; Callaghan, R.; Napier, R.M.; Bennett, M.J.; Kerr, I.D. The binding of auxin to the Arabidopsis auxin influx transporter AUX1. Plant Physiol. 2008, 148, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Peret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.; Dhondt, S.; James, N.; Casimiro, I.; Perry, P.; Syed, A.; et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.-P. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 2010, 22, 376–391. [Google Scholar] [CrossRef] [Green Version]
- Pasternak, T.; Palme, K.; Paponov, I.A. Glutathione Enhances Auxin Sensitivity in Arabidopsis Roots. Biomolecules 2020, 10, 1550. [Google Scholar] [CrossRef]
- Wang, C.L.; Oliver, D.J. Cloning of the cDNA and genomic clones for glutathione synthetase from Arabidopsis thaliana and complementation of a gsh2 mutant in fission yeast. Plant Mol. Biol. 1996, 31, 1093–1104. [Google Scholar] [CrossRef]
- Ullmann, P.; Gondet, L.; Potier, S.; Bach, T.J. Cloning of Arabidopsis thaliana glutathione synthetase (GSH2) by functional complementation of a yeast gsh2 mutant. Eur. J. Biochem. 1996, 236, 662–669. [Google Scholar] [CrossRef] [Green Version]
- Grima, G.; Benz, B.; Parpura, V.; Cuenod, M.; Do, K.Q. Dopamine-induced oxidative stress in neurons with glutathione deficit: Implication for schizophrenia. Schizophr. Res. 2003, 62, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, P.; Majhi, O.; Biswas, A.; Joshi, V.K.; Sinha, D. Enhanced accumulation of reduced glutathione by Scopoletin improves survivability of dopaminergic neurons in Parkinson’s model. Cell Death Dis. 2020, 11, 739. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Wang, Y.Q.; Mao, Q.; Wu, M.J.; Yan, Y.R.; Ren, J.J.; Wang, X.J.; Liu, A.R.; Chen, S.C. Dopamine alleviates bisphenol A-induced phytotoxicity by enhancing antioxidant and detoxification potential in cucumber. Environ. Pollut. 2020, 259, 113957. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Smalle, J.A. Auxin/cytokinin antagonistic control of the shoot/root growth ratio and its relevance for adaptation to drought and nutrient deficiency stresses. Int. J. Mol. Sci. 2022, 23, 1933. [Google Scholar] [CrossRef]
- Vernoux, T.; Wilson, R.C.; Seeley, K.A.; Reichheld, J.-P.; Muroy, S.; Brown, S.; Maughan, S.C.; Cobbett, C.S.; Van Montagu, M.; Inzé, D.; et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 2000, 12, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Koprivova, A.; Mugford, S.T.; Kopriva, S. Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep. 2010, 29, 1157–1167. [Google Scholar] [CrossRef]
- Trujillo-Hernandez, J.A.; Bariat, L.; Enders, T.A.; Strader, L.C.; Reichheld, J.P.; Belin, C. A glutathione-dependent control of the indole butyric acid pathway supports Arabidopsis root system adaptation to phosphate deprivation. J. Exp. Bot. 2020, 71, 4843–4857. [Google Scholar] [CrossRef]
- May, M.J.; Leaver, C.J. Arabidopsis thaliana gamma-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc. Natl. Acad. Sci. USA 1994, 91, 10059–10063. [Google Scholar] [CrossRef] [Green Version]
- Sági-Kazár, M.; Solymosi, K.; Solti, Á. Iron in leaves: Chemical forms, signalling, and in-cell distribution. J. Exp. Bot. 2022, 73, 1717–1734. [Google Scholar] [CrossRef]
- Jeong, J.; Connolly, E.L. Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Sci. 2009, 176, 709–714. [Google Scholar] [CrossRef]
- Shee, R.; Ghosh, S.; Khan, P.; Sahid, S.; Roy, C.; Shee, D.; Paul, S.; Datta, R. Glutathione regulates transcriptional activation of iron transporters via S-nitrosylation of bHLH factors to modulate subcellular iron homoeostasis. Plant Cell Environ. 2022, 45, 2176–2190. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Gayomba, S.R.; Jung, H.-I.; Vimalakumari, N.K.; Piñeros, M.; Craft, E.; Rutzke, M.A.; Danku, J.; Lahner, B.; Punshon, T.; et al. OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and sadmium in Arabidopsis. Plant Cell 2014, 26, 2249–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Celma, J.; Connorton, J.M.; Kruse, I.; Green, R.T.; Franceschetti, M.; Chen, Y.T.; Cui, Y.; Ling, H.Q.; Yeh, K.C.; Balk, J. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc. Natl. Acad. Sci. USA 2019, 116, 17584–17591. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, B.; Bauer, P. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 2020, 71, 1694–1705. [Google Scholar] [CrossRef] [Green Version]
- Connorton, J.M.; Balk, J.; Rodríguez-Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Shanmugam, V.; Wang, Y.-W.; Tsednee, M.; Karunakaran, K.; Yeh, K.-C. Glutathione plays an essential role in nitric oxide-mediated iron-deficiency signaling and iron-deficiency tolerance in Arabidopsis. Plant J. 2015, 84, 464–477. [Google Scholar] [CrossRef] [Green Version]
- Marchiosi, R.; Soares, A.R.; Abrahão, J.; dos Santos, W.D.; Ferrarese-Filho, O. L-DOPA and dopamine in plant metabolism. In Neurotransmitters in Plant Signaling and Communication; Baluška, F., Mukherjee, S., Ramakrishna, A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 141–167. [Google Scholar]
- Verelst, W.; Asard, H. Analysis of an Arabidopsis thaliana protein family, structurally related to cytochromes b561 and potentially involved in catecholamine biochemistry in plants. J. Plant Physiol. 2004, 161, 175–181. [Google Scholar] [CrossRef]
- Bala, K. Beyond a Neurotransmitter: Physiological Role of Dopamine in Plants. In Neurotransmitters in Plant Signaling and Communication; Baluška, F., Mukherjee, S., Ramakrishna, A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 169–187. [Google Scholar]
- Bamel, K.; Prabhavathi. Dopamine in Plant Development and Redox Signaling. In Neurotransmitters in Plant Signaling and Communication; Baluška, F., Mukherjee, S., Ramakrishna, A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 123–139. [Google Scholar]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An Auxin-Dependent Distal Organizer of Pattern and Polarity in the Arabidopsis Root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Pickett, F.B.; Wilson, A.K.; Estelle, M. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 1990, 94, 1462–1466. [Google Scholar] [CrossRef] [Green Version]
- Timpte, C.; Wilson, A.K.; Estelle, M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 1994, 138, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lee, S.; So, J.-H.; Dharmasiri, S.; Dharmasiri, N.; Ge, L.; Jensen, C.; Hangarter, R.; Hobbie, L.; Estelle, M. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 2004, 40, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Müller, B. Imaging TCSn::GFP, a synthetic cytokininreporter, in Arabidopsis thaliana. Methods Mol. Biol. 2017, 1497, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.; Guerinot, M.L. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 1996, 10, 835–844. [Google Scholar] [CrossRef]
- Song, L.; Florea, L. Rcorrector: Efficient and accurate error correction for Illumina RNA-seq reads. Gigascience 2015, 4, 48. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, V.; Contrino, S.; Cheng, C.Y.; Belyaeva, I.; Ferlanti, E.S.; Miller, J.R.; Vaughn, M.W.; Micklem, G.; Town, C.D.; Chan, A.P. ThaleMine: A warehouse for Arabidopsis data integration and discovery. Plant Cell Physiol. 2017, 58, e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurepa, J.; Smalle, J.A. Assaying transcription factor stability. In Plant Transcription Factors; Humana Press: Totowa, NJ, USA, 2011; pp. 219–234. [Google Scholar]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shull, T.E.; Kurepa, J.; Smalle, J.A. Dopamine Inhibits Arabidopsis Growth through Increased Oxidative Stress and Auxin Activity. Stresses 2023, 3, 351-371. https://doi.org/10.3390/stresses3010026
Shull TE, Kurepa J, Smalle JA. Dopamine Inhibits Arabidopsis Growth through Increased Oxidative Stress and Auxin Activity. Stresses. 2023; 3(1):351-371. https://doi.org/10.3390/stresses3010026
Chicago/Turabian StyleShull, Timothy E., Jasmina Kurepa, and Jan A. Smalle. 2023. "Dopamine Inhibits Arabidopsis Growth through Increased Oxidative Stress and Auxin Activity" Stresses 3, no. 1: 351-371. https://doi.org/10.3390/stresses3010026