Population Genetics of Chilean Jack Mackerel, Trachurus murphyi Nichols, 1920, (Pisces, Carangidae), in Waters of the South Pacific Ocean
Abstract
:1. Introduction
2. Materials and Methods
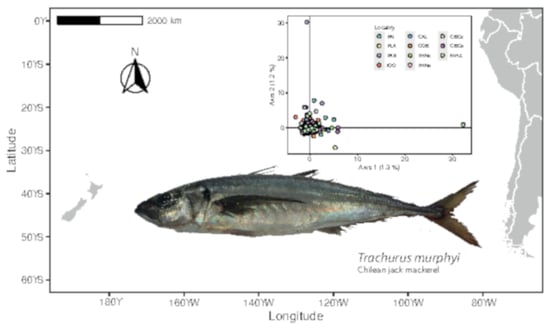
2.1. Study Area and Sample Collection
2.2. DNA Purification and PCR Procedures
2.3. Microsatellite Data Analysis
3. Results
4. Discussion
Genetic Diversity and Population Structure
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Canales-Aguirre, C.B.; Ferrada-Fuentes, S.; Galleguillos, R.; Hernández, C.E. Genetic structure in a small pelagic fish coincides with a marine protected area: Seascape genetics in Patagonian fjords. PLoS ONE 2016, 11, e0160670. [Google Scholar] [CrossRef] [Green Version]
- Galleguillos, R.; Canales-Aguirre, C.; Ferrada, S. Genetic variability in jack mackerel Trachurus murphyi Nichols: New SSRs loci and application. Gayana 2012, 76, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Canales-Aguirre, C.B.; Ferrada, S.; Hernandez, C.E.; Galleguillos, R. Population structure and demographic history of Genypterus blacodes using microsatellite loci. Fish. Res. 2010, 106, 102–106. [Google Scholar] [CrossRef]
- Valenzuela-Quiñonez, F. How fisheries management can benefit from genomics? Brief. Funct. Genomics 2016, 15, 352–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, L.; Adcock, G.J.; Smith, P.J.; Bernal Ramírez, J.H.; Carvalho, G.R. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand Snapper (Pagrus auratus). Proc. Natl. Acad. Sci. USA 2002, 99, 11742–11747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.S.; Willoughby, J.R.; Christie, M.R. Genetic diversity in fishes is influenced by habitat type and life-history variation. Ecol. Evol. 2018, 8, 12022–12031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, R. Important life history aspects of the Chilean jack mackerel, Trachurus symmetricus murphyi. Investig. Pesq. Chile 1991, 36, 67–83. [Google Scholar]
- Grechina, A. Historia de investigaciones y aspectos básicos de la ecología del jurel Trachurus symmetricus murphyi (Nichols) en alta mar del Pacífico sur. In Biología y pesca comercial del jurel en el Océano Pacífico Sur; Instituto de Investigación Pesquera: Talcahuano, Chile, 1998; pp. 11–34. [Google Scholar]
- Grechina, A.; Núñez, S.; Arcos, D. El desove del recurso jurel Trachurus symmetricus murphyi (Nichols) en el océano Pacífico sur. In Biología y pesca comercial del jurel en el Océano Pacífico Sur; Instituto de Investigación Pesquera: Talcahuano, Chile, 1998; pp. 117–140. [Google Scholar]
- Gerlotto, F.; Gutiérrez, M.; Bertrand, A. Insight on population structure of the Chilean jack mackerel (Trachurus murphyi). Aquat. Living Resour. 2012, 25, 341–355. [Google Scholar] [CrossRef] [Green Version]
- SPRFMO. 10th Scientific Committee Meeting Report; SPRFMO Publishers: Wellington, New Zealand, 2022; p. 86. [Google Scholar]
- Dioses, T.; Alarcón, V.; Nakama, M.; Echeverría, A. Desarrollo ovocitario, fecundidad parcial y distribución vertical de los cardúmenes en desove del jurel Trachurus murphyi (N); IMARPE: La Punta, Peru, 1989; pp. 287–294. [Google Scholar]
- Leal, E.; Díaz, E.; Saavedra-Nievas, J.; Claramunt, G. Ciclo reproductivo, longitud y edad de madurez del jurel Trachurus murphyi, en la costa de Chile. Rev. Biol. Mar. Oceanogr. 2013, 48, 601–611. [Google Scholar] [CrossRef] [Green Version]
- González-Kother, P.; González, M.T.; Oliva, M.E. A first assessment of atresia in the Chilean jack mackerel Trachurus murphyi (Teleostei, Carangidae) from the south-eastern Pacific Ocean. Rev. Biol. Mar. Oceanogr. 2020, 55, 100–109. [Google Scholar] [CrossRef]
- Grechina, A.; Nuñez, S.; Arcos, D. Desove del recurso jurel (Trachurus murphyi) en el océano Pacífico sur. Doc. Técn. Investig. PesqIIP Talcahuano 1994, 3, 1–44. [Google Scholar]
- Oliva, M. Metazoan parasites of the jack mackerel Trachurus murphyi (Teleostei, Carangidae) in a latitudinal gradient from South America (Chile and Peru). Parasite 1999, 6, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George-Nascimento, M. Geographical variations in the jack mackerel Trachurus symmetricus murphyi populations in the southeastern Pacific Ocean as evidenced from the associated parasite communities. J. Parasitol. 2000, 86, 929–932. [Google Scholar] [CrossRef] [PubMed]
- Cubillos, L.A.; Paramo, J.; Ruiz, P.; Núñez, S.; Sepúlveda, A. The spatial structure of the oceanic spawning of jack mackerel (Trachurus murphyi) off central Chile (1998–2001). Fish. Res. 2008, 90, 261–270. [Google Scholar] [CrossRef]
- Vásquez, S.; Correa-Ramírez, M.; Parada, C.; Sepúlveda, A. The influence of oceanographic processes on jack mackerel (Trachurus murphyi) larval distribution and population structure in the southeastern Pacific Ocean. ICES J. Mar. Sci. 2013, 70, 1097–1107. [Google Scholar] [CrossRef] [Green Version]
- Ashford, J.; Serra, R.; Saavedra, J.C.; Letelier, J. Otolith chemistry indicates large-scale connectivity in Chilean jack mackerel (Trachurus murphyi), a highly mobile species in the southern Pacific Ocean. Fish. Res. 2011, 107, 291–299. [Google Scholar] [CrossRef]
- Cárdenas, L.; Silva, A.X.; Magoulas, A.; Cabezas, J.; Poulin, E.; Ojeda, F.P. Genetic population structure in the Chilean jack mackerel, Trachurus murphyi (Nichols) across the south-eastern Pacific Ocean. Fish. Res. 2009, 100, 109–115. [Google Scholar] [CrossRef]
- Galleguillos, R.; Torres, A. Identificación de Unidades Poblacionales Pelágicas. Tercera Etapa. Informe Final; Subpesca: Valparaíso, Chile, 1988. [Google Scholar]
- González, F.; Alay, F.; Cabello, J.; Chávez, R. Definición de unidad de stock desde el punto de vista genético en el recurso jurel (Trachurus symmetricus murphyi) Nichols, 1920 (Carangidae, Perciforme). Gayana Zool. 1996, 4, 183–196. [Google Scholar]
- Sepúlveda, A.; Cubillos, L.; Grechina, A.; Peña, H.; Vilugron, L.; Hernández, A.; Miranda, L.; Sobarzo, P.; Serra, R.; Braun, M.; et al. Migración de Jurel Desde y Hacia la ZEE de Chile Central. Informes Técnicos. FIP/IT No96-15; FIP: Valparaíso, Chile, 1996; p. 273. [Google Scholar]
- Canales-Aguirre, C.B.; Ferrada, S.; Galleguillos, R. Isolation and characterization of microsatellite loci for the jack mackerel (Trachurus murphyi Nichols, 1920). Conserv. Genet. 2010, 11, 1235–1237. [Google Scholar] [CrossRef]
- Grijalva-Chon, J.; Numachi, J.; Sosa-Nishizaki, K.; de la Rosa-Vélez, J. Mitochondrial DNA Analysis of north Pacific swordfish Xiphias gladius population structure. Mar. Ecol. Prog. Ser. 1994, 115, 15–19. [Google Scholar] [CrossRef]
- Kasapidis, P.; Magoulas, A. Development and application of microsatellite markers to address the population structure of the horse mackerel Trachurus trachurus. Fish. Res. 2008, 89, 132–135. [Google Scholar] [CrossRef]
- van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Brookfield, J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996, 5, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Hedrick, P.W. Genetics of Populations, 2nd ed.; Jones and Bartlett Publishers: Boston, MA, USA, 2000. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Devillard, S.; Dufour, A.-B.; Pontier, D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 2008, 101, 92–103. [Google Scholar] [CrossRef] [Green Version]
- DeWoody, J.A.; Avise, J.C. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish. Biol. 2000, 56, 461–473. [Google Scholar] [CrossRef]
- Ward, R.D.; Woodwark, M.; Skibinski, D.O.F. A comparison of genetic diversity levels in marine, freshwater, and anadromous Fishes. J. Fish. Biol. 1994, 44, 213–232. [Google Scholar] [CrossRef]
- Canales-Aguirre, C.B.; Ferrada-Fuentes, S.; Galleguillos, R.; Oyarzun, F.X.; Hernández, C.E. Population genetic structure of Patagonian toothfish (Dissostichus eleginoides) in the southeast Pacific and southwest Atlantic Ocean. PeerJ 2018, 2018, e4173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrada, S.; Hernández, K.; Montoya, R.; Galleguillos, R. Estudio poblacional del recurso anchoveta (Engraulis ringens Jenyns 1842) (Clupeiformes, Engraulidae), mediante análisis de ADN. Gayana 2002, 66, 243–248. [Google Scholar] [CrossRef]
- Porobic, J.; Canales-Aguirre, C.B.; Ernst, B.; Galleguillos, R.; Hernandez, C.E. Biogeography and historical demography of the Juan Fernández rock lobster, Jasus frontalis (Milne Edwards, 1837). J. Hered. 2013, 104, 223–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veliz, D.; Rojas-Hernández, N.; Vega-Retter, C.; Zaviezo, C.; Garrido, I.; Pardo, L.M. Spatial and temporal stability in the genetic structure of a marine crab despite a biogeographic break. Sci. Rep. 2022, 12, 14192. [Google Scholar] [CrossRef] [PubMed]
- Veliz, D.; Rojas-Hernández, N.; Fibla, P.; Dewitte, B.; Cornejo-Guzmán, S.; Parada, C. High levels of connectivity over large distances in the Diadematid sea urchin Centrostephanus sylviae. PLoS ONE 2021, 16, e0259595. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, L.; Castilla, J.C.; Viard, F. A Phylogeographical analysis across three biogeographical provinces of the South-eastern Pacific: The case of the marine gastropod Concholepas concholepas. J. Biogeogr. 2009, 36, 969–981. [Google Scholar] [CrossRef]
- Ward, R.D. Genetics in fisheries management. Hydrobiologia 2000, 420, 191–201. [Google Scholar] [CrossRef]
- Gorbunova, N.; Evseenko, S.; Garetovsky, S. Distribution of ichthyoplankton in the frontal zones of the Peruvian waters. J. Ichthyol. 1985, 25, 67–79. [Google Scholar]
- Evseenko, S. On the reproduction of the Peruvian jack mackerel, Trachurus symmetricus murphyi (Nichols), in the southern part of the Pacific Ocean. Vopr. Ikhtiologii 1987, 27, 264–273. [Google Scholar]
- Bailey, K. Description and surface distribution of juvenile Peruvian jack mackerel, Trachurus murphyi, Nichols from the subtropical convergence zone of central south Pacific. Fish. Bull. 1989, 87, 273–278. [Google Scholar]
- Elizarov, A. Peruvian jack mackerel, Trachurus symmetricus murphyi, in the open waters of the south Pacific. J. Ichthyol 1993, 33, 86–103. [Google Scholar]
- Taylor, P. Stock structure and population biology of the Peruvian jack mackerel, Trachurus symmetricus murphyi. N. Z. Fish. Assess. Rep. 2002, 21. [Google Scholar]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Genner, M.; Collins, R.; Wellcome Sanger Institute Tree of Life Programme; Wellcome Sanger Institute Scientific Operations: DNA Pipelines Collective; Tree of Life Core Informatics Collective; Darwin Tree of Life Consortium. The genome sequence of the Atlantic horse mackerel, Trachurus trachurus (Linnaeus 1758). Wellcome Open Res. 2022, 7, 118. [Google Scholar] [CrossRef]
- Hollenbeck, C.M.; Portnoy, D.S.; Gold, J.R. Evolution of population structure in an estuarine-dependent marine Fish. Ecol. Evol. 2019, 9, 3141–3152. [Google Scholar] [CrossRef] [PubMed]
- Luikart, G.; England, P.R.; Tallmon, D.; Jordan, S.; Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat. Rev. Genet. 2003, 4, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Canales-Aguirre, C.B.; Larson, W.A.; McKinney, G.J.; Claure, C.E.; Rocha, J.D.; Ceballos, S.G.; Cádiz, M.I.; Yáñez, J.M.; Gomez-Uchida, D. Neutral and adaptive loci reveal fine-scale population structure in Eleginops maclovinus from north Patagonia. Ecol. Evol. 2022, 12, e9343. [Google Scholar] [CrossRef]


| PAI | PLA | PAR | IQQ | CAL | CQB | THNc | THNo | CBCc | CBCo | NWZ | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PAI | * | ||||||||||
| PLA | 0.002 | * | |||||||||
| PAR | 0.000 | −0.006 | * | ||||||||
| IQQ | −0.003 | −0.004 | 0.000 | * | |||||||
| CAL | 0.002 | −0.005 | −0.002 | −0.002 | * | ||||||
| CQB | 0.000 | −0.005 | 0.001 | 0.001 | 0.001 | * | |||||
| THNc | 0.003 | 0.001 | 0.000 | 0.000 | 0.000 | −0.004 | * | ||||
| THNo | 0.001 | −0.003 | −0.001 | 0.002 | 0.000 | −0.001 | 0.001 | * | |||
| CBCc | 0.000 | −0.005 | 0.001 | 0.001 | 0.000 | 0.000 | −0.001 | 0.002 | * | ||
| CBCo | 0.001 | 0.000 | 0.001 | 0.005 | 0.001 | 0.003 | 0.001 | 0.000 | 0.002 | * | |
| NWZ | 0.002 | −0.003 | 0.002 | 0.005 | −0.001 | 0.002 | 0.001 | 0.000 | 0.002 | −0.001 | * |
| Groups | FST | p-Value | FSC | p-Value | FCT | p-Value |
|---|---|---|---|---|---|---|
| Perú vs. Chile vs. New Zealand | 0.00016 | 0.665 | 0.00005 | 0.597 | 0.00011 | 0.419 |
| Panmixia | 0.00011 | 0.682 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrada Fuentes, S.; Galleguillos, R.; Herrera-Yáñez, V.; Canales-Aguirre, C.B. Population Genetics of Chilean Jack Mackerel, Trachurus murphyi Nichols, 1920, (Pisces, Carangidae), in Waters of the South Pacific Ocean. Fishes 2023, 8, 162. https://doi.org/10.3390/fishes8030162
Ferrada Fuentes S, Galleguillos R, Herrera-Yáñez V, Canales-Aguirre CB. Population Genetics of Chilean Jack Mackerel, Trachurus murphyi Nichols, 1920, (Pisces, Carangidae), in Waters of the South Pacific Ocean. Fishes. 2023; 8(3):162. https://doi.org/10.3390/fishes8030162
Chicago/Turabian StyleFerrada Fuentes, Sandra, Ricardo Galleguillos, Victoria Herrera-Yáñez, and Cristian B. Canales-Aguirre. 2023. "Population Genetics of Chilean Jack Mackerel, Trachurus murphyi Nichols, 1920, (Pisces, Carangidae), in Waters of the South Pacific Ocean" Fishes 8, no. 3: 162. https://doi.org/10.3390/fishes8030162