Root Physiological Changes and Transcription Analysis of Iris domestica in Response to Persistent Drought
Abstract
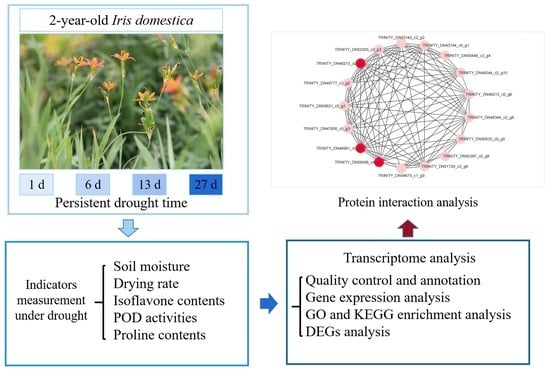
:1. Introduction
2. Materials and Methods
3. Results
3.1. Soil Water Content and Root Physiological Characteristics
3.2. Quality Control and Annotation
3.3. Gene Expression Analysis
3.4. GO and KEGG Enrichment Analysis
3.5. DEGs Related to Water Deprivation and Oxidative Stress
3.6. Expression of Genes Involved in Metabolism and Biosynthesis
3.7. Transcription Factors Responding to Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kleinwächter, M.; Selmar, D. Influencing the product quality by applying drought stress during the cultivation of medicinal plants. In Physiological Mechanisms and Adaptation Strategies in Plants under Changing Environment; Springer: Berlin/Heidelberg, Germany, 2014; pp. 57–73. [Google Scholar]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Pant, P.; Pandey, S.; Dall’Acqua, S. The influence of environmental conditions on secondary metabolites in medicinal plants: A literature review. Chem. Biodivers. 2021, 18, e2100345. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.M.; Percival, G.C.; Rose, G. Variations in seasonal drought tolerance rankings. Trees 2019, 33, 1063–1072. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Xia, H.; Lu, B. Crop breeding for drought resistance. Front. Plant Sci. 2019, 10, 314. [Google Scholar] [CrossRef]
- Li, P.; Zhu, Y.; Song, X.; Song, F. Negative effects of long-term moderate salinity and short-term drought stress on the photosynthetic performance of Hybrid Pennisetum. Plant Physiol. Biochem. 2020, 155, 93–104. [Google Scholar] [CrossRef]
- Ors, S.; Ekinci, M.; Yildirim, E.; Sahin, U.; Turan, M.; Dursun, A. Interactive effects of salinity and drought stress on photosynthetic characteristics and physiology of tomato (Lycopersicon esculentum L.) seedlings. S. Afr. J. Bot. 2021, 137, 335–339. [Google Scholar] [CrossRef]
- Widuri, L.I.; Lakitan, B.; Sakagami, J.; Yabuta, S.; Kartika, K.; Siaga, E. Short-term drought exposure decelerated growth and photosynthetic activities in chili pepper (Capsicum annuum L.). Ann. Agric. Sci. 2020, 65, 149–158. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Han, M.; Yang, L.; Li, Y.; Sun, Z.; Zhang, T. Changes in the physiological characteristics and baicalin biosynthesis metabolism of Scutellaria baicalensis Georgi under drought stress. Ind. Crop. Prod. 2018, 122, 473–482. [Google Scholar] [CrossRef]
- Bozin, B.; Mimica-Dukic, N.; Samojlik, I.; Goran, A.; Igic, R. Phenolics as antioxidants in garlic (Allium sativum L., Alliaceae). Food Chem. 2008, 111, 925–929. [Google Scholar] [CrossRef]
- Mittler, R.; Zilinskas, B.A. Regulation of pea cytosolic ascorbate peroxidase and other antioxidant enzymes during the progression of drought stress and following recovery from drought. Plant J. 1994, 5, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, L.; Yang, X.; Zhang, T.; Lan, Y.; Zhao, Y.; Han, M.; Yang, L. Drought stress induces biosynthesis of flavonoids in leaves and saikosaponins in roots of Bupleurum chinense DC. Phytochemistry 2020, 177, 112434. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.J.; Weon, S.Y. Effect of progressive drought stress on physio-biochemical responses and gene expression patterns in wheat. 3 Biotech 2021, 11, 440. [Google Scholar]
- He, C.; Du, Y.; Fu, J.; Zeng, E.; Park, S.; White, F.; Zheng, J.; Liu, S. Early Drought-Responsive genes are variable and relevant to drought tolerance. G3 2020, 10, 1657–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Qu, J.; Guo, X.; Li, L.; Zhang, X.; Yang, Q.; Lu, Y.; Li, W.; Fu, F. Overexpression of vacuolar H+-pyrophosphatase (H+-PPase) gene from Ammopiptanthus nanus enhances drought tolerance in maize. J. Agron. Crop Sci. 2021, 208, 633–644. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Xie, Z.; Guo, D.; Chen, C.; Fan, Q.; Deng, X.; Liu, J.H. Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata. Plant Biotechnol. J. 2019, 17, 1394–1407. [Google Scholar] [CrossRef] [Green Version]
- Aduse Poku, S.; Nkachukwu Chukwurah, P.; Aung, H.H.; Nakamura, I. Over-expression of a melon Y3SK2-type LEA gene confers drought and salt tolerance in transgenic tobacco plants. Plants 2020, 9, 1749. [Google Scholar] [CrossRef]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol. Progr. 2011, 27, 297–306. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Shi, J.; Wu, M.; Xu, D. The application and future prospect of RNA-Seq technology in Chinese medicinal plants. J. Appl. Res. Med. Aromat. Plants 2021, 24, 100318. [Google Scholar] [CrossRef]
- Anderson, N.O. Breeding for dwarf, winter-hardy Iris domestica, blackberry lily (Iridaceae). Acta Hortic 2018, 1263, 275–282. [Google Scholar] [CrossRef]
- Yang, X.; Guo, S.; Feng, M.; Lai, X.; Shang, J. Effects of drought stress and re-watering on the characteristics of photosynthesis and chlorophyll fluorescence of blackberry lily. Acta Agric. Univ. Jiangxiensis 2018, 40, 525–532. [Google Scholar]
- Zhu, Y.; Pu, B.Q.; Xie, G.Y.; Tian, M.; Xu, F.Y.; Qin, M.J. Dynamic changes of flavonoids contents in the different parts of rhizome of Belamcanda chinensis during the thermal drying process. Molecules 2014, 19, 10440–10454. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bertini, L.; Proietti, S.; Focaracci, F.; Canini, F.; Bravo, L.A.; Rabert, C.; Caruso, C. Identification and validation of new reference genes for accurate quantitative reverse transcriptase-PCR normalization in the Antarctic plant Colobanthus quitensis under abiotic stress conditions. Polar Biol. 2021, 44, 389–405. [Google Scholar] [CrossRef]
- Ai, Q.; Liu, C.; Han, M.; Yang, L. Selection and Verification of Reference Genes for qRT-PCR Analysis in Iris domestica under Drought. Phyton-Int. J. Exp. Bot. 2022, 91, 2537–2548. [Google Scholar] [CrossRef]
- Zlatev, Z.; Lidon, F.C. An overview on drought induced changes in plant growth, water relations and photosynthesis. Emir. J. Food Agric. 2012, 24, 57. [Google Scholar]
- Wang, S.; Liang, D.; Li, C.; Hao, Y.; Ma, F.; Shu, H. Influence of drought stress on the cellular ultrastructure and antioxidant system in leaves of drought-tolerant and drought-sensitive apple rootstocks. Plant Physiol. Biochem. 2012, 51, 81–89. [Google Scholar] [CrossRef]
- Terzi, R.A.A.K. Drought stress tolerance and the antioxidant enzyme system. Acta Biol. Cracov. Bot. 2006, 48, 89–96. [Google Scholar]
- Yang, L.; Zhao, Y.; Zhang, Q.; Cheng, L.; Han, M.; Ren, Y.; Yang, L. Effects of drought-re-watering-drought on the photosynthesis physiology and secondary metabolite production of Bupleurum chinense DC. Plant Cell Rep. 2019, 38, 1181–1197. [Google Scholar] [CrossRef]
- Huang, L.Q.; Guo, L.P. Secondary metabolites accumulating and geoherbs formation under enviromental stress. China J. Chin. Mater. Med. 2007, 32, 277–280. [Google Scholar]
- Al-Gabbiesh, A.; Selmar, D.; Kleinwächter, M. Influencing the contents of secondary metabolites in spice and medicinal plants by deliberately applying drought stress during their cultivation. Jordan J. Biol. Sci. 2015, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Wang, Y.; Yu, S.; Huang, Y.; Liu, H.; Chen, W.; He, X. Effects of drought stress on growth, physiology and secondary metabolites of Two Adonis species in Northeast China. Sci. Hortic.-Amst. 2020, 259, 108795. [Google Scholar] [CrossRef]
- Xin, J.; Sun, C.; Li, G.; Li, G.; Chen, G. Effects of progressive drought stress on the physiology, antioxidative enzymes and secondary metabolites of Radix Astragali. Acta Physiol. Plant. 2015, 37, 262. [Google Scholar]
- Qin, M.J.; Liang, J.W.; Liu, J.; Zhao, J.; Dian, Y.G. Scavenging effects on radicals of isoflavones from rhizome of Belamcandae Chinensis. Chin. Tradit. Herb. Drugs 2003, 34, 640–641. [Google Scholar]
- Xie, G.Y.; Zhu, Y.; Shu, P.; Qin, X.Y.; Wu, G.; Wang, Q.; Qin, M.J. Phenolic metabolite profiles and antioxidants assay of three Iridaceae medicinal plants for traditional Chinese medicine “She-gan” by on-line HPLC–DAD coupled with chemiluminescence (CL) and ESI-Q-TOF-MS/MS. J. Pharmaceut. Biomed. 2014, 98, 40–51. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y.; Ye, J.; Ye, Z.; Zhu, R.; Xie, G.; Zhao, Y.; Qin, M. Iris domestica (iso) flavone 7-and 3′-O-Glycosyltransferases Can be Induced by CuCl2. Front. Plant Sci. 2021, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H. Anesthetic agents of plant origin: A review of phytochemicals with anesthetic activity. Molecules 2017, 22, 1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.Q.; Han, N.; Zhang, Y.Y.; Tao, Y.; Chen, L.; Liu, Y.P.; Zhou, S.F.; Fu, F.L.; Li, W.C. Cloning and characterization of vacuolar H+-pyrophosphatase gene (AnVP1) from Ammopiptanthus nanus and its heterologous expression enhances osmotic tolerance in yeast and Arabidopsis thaliana. Plant Growth Regul. 2017, 81, 385–397. [Google Scholar] [CrossRef]
- Su, J.H.; Bai, T.H.; Wang, F.; Bao, A.K. Overexpression of Arabidopsis H+-pyrophosphatase improves the growth of alfalfa under long-term salinity, drought conditions and phosphate deficiency. Czech J. Genet. Plant 2019, 55, 156–161. [Google Scholar] [CrossRef]
- Colmenero-Flores, J.M.; Campos, F.; Garciarrubio, A.; Covarrubias, A.A. Characterization of Phaseolus vulgaris cDNA clones responsive to water deficit: Identification of a novel late embryogenesis abundant-like protein. Plant Mol. Biol. 1997, 35, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Lu, H.; Zhou, Q.; Xie, H.T.; Li, J.Y.; Chen, Z.Y.; Han, S.C.; Zhao, H.P.; Zhao, H.X. Analysis of LEA protein family members in Lepidium apetalum seeds and the expression of LaLEA1 in seedlings in response to abiotic stresses. Biol. Plant. 2020, 64, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Liu, Q.; Xu, W.; Zhang, J.; Wang, X.; Nie, G.; Yao, L.; Wang, H.; Lin, C. De novo assembly and discovery of genes that involved in drought tolerance in the common vetch. Int. J. Mol. Sci. 2019, 20, 328. [Google Scholar] [CrossRef]
- Wang, R.; Wu, H.; Zhang, M.; Ni, Z.; Sun, Q. Cloning, characterization and transgenic function analysis of wheat (Triticum aestivum L.) TaWRKY51 gene. J. Agric. Biotechnol. 2013, 21, 1019–1027. [Google Scholar]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ai, Q.; Sun, Y.; Dai, A.; LYU, Z.; Liu, C.; Han, M.; Yang, L. Root Physiological Changes and Transcription Analysis of Iris domestica in Response to Persistent Drought. Horticulturae 2022, 8, 1162. https://doi.org/10.3390/horticulturae8121162
Ai Q, Sun Y, Dai A, LYU Z, Liu C, Han M, Yang L. Root Physiological Changes and Transcription Analysis of Iris domestica in Response to Persistent Drought. Horticulturae. 2022; 8(12):1162. https://doi.org/10.3390/horticulturae8121162
Chicago/Turabian StyleAi, Qiang, Yingxin Sun, Ailin Dai, Zeliang LYU, Cuijing Liu, Mei Han, and Limin Yang. 2022. "Root Physiological Changes and Transcription Analysis of Iris domestica in Response to Persistent Drought" Horticulturae 8, no. 12: 1162. https://doi.org/10.3390/horticulturae8121162