Effects of Four Critical Gene Deletions in Saccharomyces cerevisiae on Fusel Alcohols during Red Wine Fermentation
Abstract
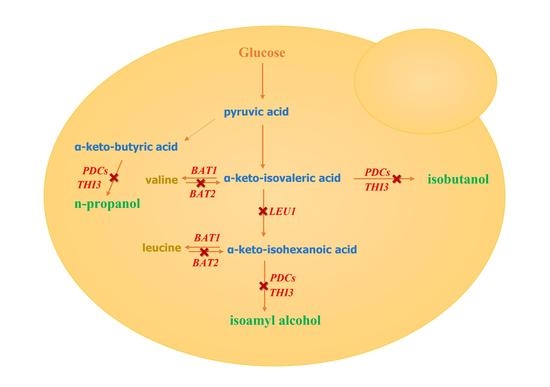
:1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Design and Synthesis of Primers
2.3. Construction of the Disruption Cassette
2.4. Yeast Transformation and Screening
2.5. Construction of the Diploid S. cerevisiae
2.6. Real-Time Quantitative PCR
2.7. Fermentation Analysis of Recombinant Yeasts in Red Wine Fermentation
2.8. Analytical Methods
2.9. Statistical Analysis
3. Results
3.1. Construction of Recombinant Yeast Strains
3.2. Relative Expression Levels of LEU1, PDC5, and PDC1
3.3. Comparison of Fermentation Performance of Different Recombinant Strains
3.4. Effects of BAT2, THI3, and PDC5 Deletion on the Production of Fusel Alcohols
3.5. Effect of LEU1 Deletion on the Production of Fusel Alcohols
3.6. Comparison of Total Fusel Alcohols of Different Recombinant Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, K.J.; Liz Thach, M.; Olsen, J. Wine and health perceptions: Exploring the impact of gender, age and ethnicity on consumer perceptions of wine and health. Wine Econ. Policy 2016, 5, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Huang, S.; Du, L.; Tang, P.; Xiao, D. Reduced Production of Higher Alcohols by Saccharomyces cerevisiae in Red Wine Fermentation by Simultaneously Overexpressing BAT1 and Deleting BAT2. J. Agric. Food Chem. 2017, 65, 6936–6942. [Google Scholar] [CrossRef] [PubMed]
- Connell, D.W.; Strauss, C.R. Major constituents of fusel oils distilled from Australian grape wines. J. Sci. Food Agric. 1974, 25, 31–44. [Google Scholar] [CrossRef]
- Xie, J.; Tian, X.; He, S.; Wei, Y.; Peng, B.; Wu, Z. Evaluating the Intoxicating Degree of Liquor Products with Combinations of Fusel Alcohols, Acids, and Esters. Molecules 2018, 23, 1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Zeng, X. Effect of activated carbon on bitterness substances of litchi spirit. Sci. Technol. Food Ind. 2012, 4, 291–293. [Google Scholar]
- Luo, Y.; Kong, L.; Xue, R.; Wang, W.; Xia, X. Bitterness in alcoholic beverages: The profiles of perception, constituents, and contributors. Trends Food Sci. Technol. 2020, 96, 222–232. [Google Scholar] [CrossRef]
- Qi, X.; Shi, X.; Wang, J.; Ma, Y. Determination of Methanol, Ethyl Acetate and Higher Alcohols Content in Cabernet Sauvignon Dry Red Wine. Liquor-Mak. Sci. Technol. 2018, 3, 98–101. [Google Scholar] [CrossRef]
- Eden, A.; Van Nedervelde, L.; Drukker, M.; Benvenisty, N.; Debourg, A. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl. Microbiol. Biotechnol. 2001, 55, 296–300. [Google Scholar] [CrossRef]
- Hazelwood, L.A.; Daran, J.M.; van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Liu, L.; Wang, Y.; Wang, X.; Xiao, D. Higher alcohols metabolism by Saccharomyces cerevisiae: A mini review. Sheng Wu Gong Cheng Xue Bao 2021, 37, 429–447. [Google Scholar] [CrossRef]
- Colón, M.; Hernández, F.; López, K.; Quezada, H.; González, J.; López, G.; Aranda, C.; González, A. Saccharomyces cerevisiae Bat1 and Bat2 Aminotransferases Have Functionally Diverged from the Ancestral-Like Kluyveromyces lactis Orthologous Enzyme. PLoS ONE 2011, 6, e16099. [Google Scholar] [CrossRef] [Green Version]
- Styger, G.; Jacobson, D.; Prior, B.A.; Bauer, F.F. Genetic analysis of the metabolic pathways responsible for aroma metabolite production by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2013, 97, 4429–4442. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, J.R.; Norte, V. A study of branched-chain amino acid aminotransferase and isolation of mutations affecting the catabolism of branched-chain amino acids in Saccharomyces cerevisiae. FEBS Lett. 1993, 326, 29–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skala, J.; Capieaux, E.; Balzi, E.; Chen, W.N.; Goffeau, A. Complete sequence of the Saccharomyces cerevisiae LEU1 gene encoding isopropylmalate isomerase. Yeast 1991, 7, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, S.; Wang, J.; Zhang, C.; Shi, Y.; Guo, X.; Chen, Y.; Xiao, D. Genetic engineering to alter carbon flux for various higher alcohol productions by Saccharomyces cerevisiae for Chinese Baijiu fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, J.R.; Harrison, S.J.; Hewlins, M.J. An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 25751–25756. [Google Scholar] [CrossRef] [Green Version]
- Romagnoli, G.; Luttik, M.A.H.; Kötter, P.; Pronk, J.T.; Daran, J.M. Substrate specificity of thiamine pyrophosphate-dependent 2-oxo-acid decarboxylases in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 7538–7548. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, J.R.; Salgado, L.E.J.; Hewlins, M.J.E. The Catabolism of Amino Acids to Long Chain and Complex Alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 8028–8034. [Google Scholar] [CrossRef] [Green Version]
- Hao, X.; Xiao, D.; Zhang, C. Effect of YDL080C gene deletion on higher alcohols production in Saccharomyces cerevisiae haploids. Wei Sheng Wu Xue Bao = Acta Microbiol. Sin. 2010, 50, 1030–1035. [Google Scholar]
- Li, T.; Sun, J.; Wu, D.; Li, X.M.; Xie, G.F.; Lu, J. Effect of YDL080C and LEU2 gene knockout on isoamyl alcohol production in industrial yellow rice wine yeast. Sci. Technol. Food Ind. 2015, 36, 1891193. [Google Scholar]
- Hohmann, S.; Cederberg, H. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur. J. Biochem. 1990, 188, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. PDC6, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when fused spontaneously under the control of the PDC1 promoter. Curr. Genet. 1991, 20, 373–378. [Google Scholar] [CrossRef]
- Wu, L.; Wen, Y.; Chen, W.; Yan, T.; Tian, X.; Zhou, S. Simultaneously deleting ADH2 and THI3 genes of Saccharomyces cerevisiae for reducing the yield of acetaldehyde and fusel alcohols. FEMS Microbiol. Lett. 2021, 368, fnab094. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, S.; Yan, T.; Liang, S. Construction of THI3/BAT2 Gene-Deleted Saccharomyces cerevisiae and Its Application in Preparing Chinese Rice Wine. Mod. Food Sci. Technol. 2022, 11, 55–62. [Google Scholar]
- Wang, Y.; Sun, Z.; Zhang, C.; Zhang, Q.; Guo, X.; Xiao, D. Comparative transcriptome analysis reveals the key regulatory genes for higher alcohol formation by yeast at different α-amino nitrogen concentrations. Food Microbiol. 2021, 95, 103713. [Google Scholar] [CrossRef] [PubMed]
- Mataffo, A.; Scognamiglio, P.; Dente, A.; Strollo, D.; Colla, G.; Rouphael, Y.; Basile, B. Foliar Application of an Amino Acid-Enriched Urea Fertilizer on ‘Greco’ Grapevines at Full Veraison Increases Berry Yeast-Assimilable Nitrogen Content. Plants 2020, 9, 619. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.L.; Budde, A.D. Effect of Reducing and Oxidizing Agents and pH on Malt Endoproteolytic Activities and Brewing Mashes. J. Agric. Food Chem. 2003, 51, 7504–7512. [Google Scholar] [CrossRef]
- Okuda, M.; Miyamoto, M.; Joyo, M.; Takahashi, K.; Goto-Yamamoto, N.; Iida, S.; Ishii, T. The relationship between rice protein composition and nitrogen compounds in sake. J. Biosci. Bioeng. 2016, 122, 70–78. [Google Scholar] [CrossRef]
- González, C.; Perdomo, G.; Tejera, P.; Brito, N.; Siverio, J.M. One-step, PCR-mediated, gene disruption in the yeast Hansenula polymorpha. Yeast 1999, 15, 1323–1329. [Google Scholar] [CrossRef]
- Meilhoc, E.; Teissie, J. Electrotransformation of Saccharomyces cerevisiae. In Electroporation Protocols: Microorganism, Mammalian System, and Nanodevice; Li, S., Chang, L., Teissie, J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 187–193. [Google Scholar]
- Azevedo, F.; Pereira, H.; Johansson, B. Colony PCR. In PCR: Methods and Protocols; Domingues, L., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 129–139. [Google Scholar]
- Güldener, U.; Heck, S.; Fiedler, T.; Beinhauer, J.; Hegemann, J.H. A New Efficient Gene Disruption Cassette for Repeated Use in Budding Yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef] [Green Version]
- de-la-Fuente-Blanco, A.; Sáenz-Navajas, M.P.; Ferreira, V. Levels of higher alcohols inducing aroma changes and modulating experts’ preferences in wine model solutions. Aust. J. Grape Wine Res. 2017, 23, 162–169. [Google Scholar] [CrossRef]
- Sun, Z.; Zhou, B.; Wang, M.; Wang, Y.; Xing, S.; Guo, X.; Xiao, D. Construction of industrial brewing yeast for fermentation under high temperature and high gravity condition. Sheng Wu Gong Cheng Xue Bao 2019, 35, 522–534. [Google Scholar] [PubMed]
- Brion, C.; Ambroset, C.; Delobel, P.; Sanchez, I.; Blondin, B. Deciphering regulatory variation of THI genes in alcoholic fermentation indicate an impact of Thi3p on PDC1 expression. BMC Genom. 2014, 15, 1085. [Google Scholar] [CrossRef] [Green Version]
- Styger, G.; Jacobson, D.; Bauer, F.F. Identifying genes that impact on aroma profiles produced by Saccharomyces cerevisiae and the production of higher alcohols. Appl. Microbiol. Biotechnol. 2011, 91, 713–730. [Google Scholar] [CrossRef] [Green Version]
- Lilly, M.; Bauer, F.F.; Styger, G.; Lambrechts, M.G.; Pretorius, I.S. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res. 2006, 6, 726–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.P.; Schimmel, P. Yeast LEU1. Repression of mRNA levels by leucine and relationship of 5′-noncoding region to that of LEU2. J. Biol. Chem. 1984, 259, 3714–3719. [Google Scholar] [CrossRef]
- Mangas, J.J.; Cabranes, C.; Moreno, J.; Gomis, D.B. Influence of Cider-Making Technology on Cider Taste. LWT Food Sci. Technol. 1994, 27, 583–586. [Google Scholar] [CrossRef] [Green Version]




| Strains | Ethanol (%vol) | pH Value | Residual Sugar (g·L−1) | Total Acid (g·L−1) |
|---|---|---|---|---|
| XF1 | 10.43 ± 0.21 | 3.28 ± 0.01 | 0.77 ± 0.11 | 8.63 ± 0.24 |
| XF-T | 10.45 ± 0.26 | 3.27 ± 0.02 | 0.83 ± 0.08 | 8.75 ± 0.18 |
| XF-B | 10.98 ± 0.33 | 3.30 ± 0.01 | 0.82 ± 0.13 | 8.63 ± 0.27 |
| XF-P | 10.64 ± 0.18 | 3.28 ± 0.01 | 0.87 ± 0.19 | 9.00 ± 0.33 |
| XF-L | 10.97 ± 0.25 | 3.27 ± 0.01 | 0.79 ± 0.15 | 8.75 ± 0.13 |
| XF-TP | 10.78 ± 0.17 | 3.32 ± 0.01 | 0.78 ± 0.06 | 8.38 ± 0.29 |
| XF-TB | 10.76 ± 0.24 | 3.31 ± 0.02 | 0.83 ± 0.17 | 8.63 ± 0.15 |
| XF-BP | 10.75 ± 0.22 | 3.32 ± 0.01 | 0.86 ± 0.12 | 8.50 ± 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, T.; Wang, Z.; Zhou, H.; He, J.; Zhou, S. Effects of Four Critical Gene Deletions in Saccharomyces cerevisiae on Fusel Alcohols during Red Wine Fermentation. Fermentation 2023, 9, 379. https://doi.org/10.3390/fermentation9040379
Yan T, Wang Z, Zhou H, He J, Zhou S. Effects of Four Critical Gene Deletions in Saccharomyces cerevisiae on Fusel Alcohols during Red Wine Fermentation. Fermentation. 2023; 9(4):379. https://doi.org/10.3390/fermentation9040379
Chicago/Turabian StyleYan, Tongshuai, Zexiang Wang, Haoyang Zhou, Jiaojiao He, and Shishui Zhou. 2023. "Effects of Four Critical Gene Deletions in Saccharomyces cerevisiae on Fusel Alcohols during Red Wine Fermentation" Fermentation 9, no. 4: 379. https://doi.org/10.3390/fermentation9040379