Diagnostic and Prognostic Value of microRNAs in Patients with Laryngeal Cancer: A Systematic Review
Abstract
:1. Introduction
2. Results
2.1. Study Selection
2.2. Characteristics of the Selected Studies
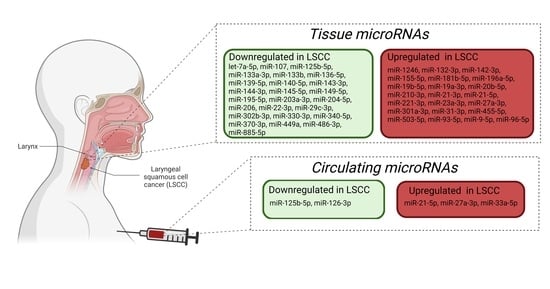
2.3. miRNA and LSCC
2.4. Diagnostic Potential of miRNA in LSCC
2.5. Prognostic Potential of miRNA in LSCC
2.6. Functional Role of miRNA in Laryngeal Squamous Cell Carcinoma
3. Discussion
4. Materials and Methods
4.1. Search Strategy
4.2. Inclusion and Exclusion Criteria
4.3. Data Extraction
4.4. Study Quality Assessment
4.5. Statistical Analysis
4.6. miRNA Nomenclature
4.7. Study Selection of Functional Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, S.; Mandal, S.; Banerjee, S.; Mandal, G.K.; Bhowmick, A.K.; Murmu, N. Cannabis smoke can be a major risk factor for early-age laryngeal cancer—A molecular signaling-based approach. Tumor Biol. 2015, 36, 6029–6036. [Google Scholar] [CrossRef] [PubMed]
- Mourad, M.; Jetmore, T.; Jategaonkar, A.A.; Moubayed, S.; Moshier, E.; Urken, M.L. Epidemiological Trends of Head and Neck Cancer in the United States: A SEER Population Study. J. Oral Maxillofac. Surg. 2017, 75, 2562–2572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machiels, J.-P.; Leemans, C.R.; Golusinski, W.; Grau, C.; Licitra, L.; Gregoire, V. Reprint of “Squamous cell carcinoma of the oral cavity, larynx, oropharynx and hypopharynx: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up”. Oral Oncol. 2020, 113, 105042. [Google Scholar] [CrossRef] [PubMed]
- Brandstorp-Boesen, J.; Falk, R.S.; Evensen, J.F.; Boysen, M.; Brøndbo, K. Risk of Recurrence in Laryngeal Cancer. PLoS ONE 2016, 11, e0164068. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [Green Version]
- Fu, G.; Brkić, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [Green Version]
- Tüfekci, K.U.; Öner, M.G.; Meuwissen, R.L.J.; Genç, Ş. The Role of MicroRNAs in Human Diseases. Methods Mol. Biol. 2013, 1107, 33–50. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Negrini, M.; Ferracin, M.; Sabbioni, S.; Croce, C.M. MicroRNAs in human cancer: From research to therapy. J. Cell Sci. 2007, 120, 1833–1840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riefolo, M.; Porcellini, E.; Dika, E.; Broseghini, E.; Ferracin, M. Interplay between small and long non-coding RNA s in cutaneous melanoma: A complex jigsaw puzzle with missing pieces. Mol. Oncol. 2019, 13, 74–98. [Google Scholar] [CrossRef] [PubMed]
- Durante, G.; Comito, F.; Lambertini, M.; Broseghini, E.; Dika, E.; Ferracin, M. Non-coding RNA dysregulation in skin cancers. Essays Biochem. 2021, 65, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.; Anderson, T. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar]
- Ferracin, M.; Negrini, M. Micromarkers 2.0: An update on the role of microRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 2015, 15, 1369–1381. [Google Scholar] [CrossRef]
- Cocks, A.; Martinez-Rodriguez, V.; Del Vecchio, F.; Schukking, M.; Broseghini, E.; Giannakopoulos, S.; Fabbri, M. Diverse roles of EV-RNA in cancer progression. Semin. Cancer Biol. 2021, 75, 127–135. [Google Scholar] [CrossRef]
- Durante, G.; Broseghini, E.; Comito, F.; Naddeo, M.; Milani, M.; Salamon, I.; Campione, E.; Dika, E.; Ferracin, M. Circulating microRNA biomarkers in melanoma and non-melanoma skin cancer. Expert Rev. Mol. Diagn. 2022, 22, 305–318. [Google Scholar] [CrossRef]
- Kang, J.-W.; Eun, Y.-G.; Lee, Y.-C. Diagnostic Value of Salivary miRNA in Head and Neck Squamous Cell Cancer: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2021, 22, 7026. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, Z.; Zhao, J.; Zhai, X.; Li, J.; Zhang, Y.; Zong, L.; Peng, H.; Qi, J.; Kong, X.; et al. H19/miR-107/HMGB1 axis sensitizes laryngeal squamous cell carcinoma to cisplatin by suppressing autophagy in vitro and in vivo. Cell Biol. Int. 2021, 45, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.-M.; Yang, H.; Yuan, Z.-N.; Yang, X.-G.; Pei, R.; He, H.-J. Long noncoding RNA LINC01194 enhances the malignancy of laryngeal squamous cell carcinoma by sponging miR-655 to increase SOX18 expression. Biochem. Biophys. Res. Commun. 2020, 529, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Wu, S.; Hu, H.; Dai, X.; Wang, X. Long Noncoding RNA MIAT Regulates the Process of Laryngeal Squamous Cell Carcinoma Through Regulation of miR-147a/BCOR. Arch. Med. Res. 2021, 52, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, J.; Shan, C.; Zhang, H.; Xu, O. MicroRNA-892a regulates laryngocarcinoma cell proliferation via Dicer. Exp. Biol. Med. 2020, 245, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Tuncturk, F.R.; Akalin, I.; Uzun, L.; Zenginkinet, T. Comparison of miRNA expressions among benign, premalignant and malignant lesions of the larynx: Could they be transformation biomarkers? J. Otolaryngol. Head Neck Surg. 2021, 50, 14. [Google Scholar] [CrossRef]
- He, G.; Pang, R.; Han, J.; Jia, J.; Ding, Z.; Bi, W.; Yu, J.; Chen, L.; Zhang, J.; Sun, Y. TINCR inhibits the proliferation and invasion of laryngeal squamous cell carcinoma by regulating miR-210/BTG2. BMC Cancer 2021, 21, 753. [Google Scholar] [CrossRef]
- Kawasaki, H.; Takeuchi, T.; Ricciardiello, F.; Lombardi, A.; Biganzoli, E.; Fornili, M.; De Bortoli, D.; Mesolella, M.; Cossu, A.M.; Scrima, M.; et al. Definition of miRNA Signatures of Nodal Metastasis in LCa: miR-449a Targets Notch Genes and Sup-presses Cell Migration and Invasion. Mol. Ther. Nucleic Acids 2020, 20, 711–724. [Google Scholar] [CrossRef]
- Cao, J.; Yang, Z.; An, R.; Zhang, J.; Zhao, R.; Li, W.; Xu, L.; Sun, Y.; Liu, M.; Tian, L. lncRNA IGKJ2-MALLP2 suppresses LSCC proliferation, migration, invasion, and angiogenesis by sponging miR-1911-3p/p21. Cancer Sci. 2020, 111, 3245–3257. [Google Scholar] [CrossRef]
- Song, K.; Yu, P.; Zhang, C.; Yuan, Z.; Zhang, H. The LncRNA FGD5-AS1/miR-497-5p axis regulates septin 2 (SEPT2) to accelerate cancer progression and increase cisplatin-resistance in laryngeal squamous cell carcinoma. Mol. Carcinog 2021, 60, 469–480. [Google Scholar] [CrossRef]
- Pantazis, T.-L.; Giotakis, A.I.; Karamagkiolas, S.; Giotakis, I.; Konstantoulakis, M.; Liakea, A.; Misiakos, E.P. Low expression of miR-20b-5p indicates favorable prognosis in laryngeal squamous cell carcinoma, especially in patients with non-infiltrated regional lymph nodes. Am. J. Otolaryngol. 2020, 41, 102563. [Google Scholar] [CrossRef] [PubMed]
- Popov, T.M.; Giragosyan, S.; Petkova, V.; Stancheva, G.; Marinov, T.; Belitova, M.; Rangachev, J.; Popova, D.; Kaneva, R.P. Proangiogenic signature in advanced laryngeal carcinoma after microRNA expression profiling. Mol. Biol. Rep. 2020, 47, 5651–5655. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zheng, X.; Guo, H.; Xue, X.; Zhang, Y.; Niu, M.; Cui, J.; Liu, H.; Luo, H.; Yang, D.; et al. Serum Exosomal miR-941 as a promising Oncogenic Biomarker for Laryngeal Squamous Cell Carcinoma. J. Cancer 2020, 11, 5329–5344. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Zheng, X.; Dai, F.; Lu, Y.; Dai, L.; Niu, M.; Guo, H.; Li, W.; Xue, X.; et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol. Cancer 2020, 19, 99. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Chen, W.; Li, C.; Chen, X.; Lin, Q.; Lin, S.; Wang, D. Circular RNA circ_0004507 contributes to laryngeal cancer progression and cisplatin resistance by sponging miR -873 to upregulate multidrug resistance 1 and multidrug resistance protein 1. Head Neck 2021, 43, 928–941. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, Z.; Yu, G. microRNA-139-3p Inhibits Malignant Behaviors of Laryngeal Cancer Cells via the KDM5B/SOX2 Axis and the Wnt/beta-Catenin Pathway. Cancer Manag. Res. 2020, 12, 9197–9209. [Google Scholar] [CrossRef]
- Zheng, Y.; Duan, L.; Yang, Y.; Luo, D.; Yan, B. Circ_0023028 contributes to the progression of laryngeal squamous cell carcinoma by upregulating LASP1 through miR-486-3p. Mol. Cell. Biochem. 2021, 476, 2951–2961. [Google Scholar] [CrossRef]
- Shuang, Y.; Liu, J.; Niu, J.; Guo, W.; Li, C. A novel circular RNA circPPFIA1 promotes laryngeal squamous cell carcinoma progression through sponging miR-340-3p and regulating ELK1 expression. Bioengineered 2021, 12, 5220–5230. [Google Scholar] [CrossRef]
- Kyurkchiyan, S.G.; Popov, T.M.; Shakola, F.; Rangachev, J.; Mitev, V.I.; Kaneva, R. A Pilot Study Reveals the Potential of miR-31-3p and miR-196a-5p as Non-Invasive Biomarkers in Advanced Laryngeal Cancer. Folia Medica 2021, 63, 355–364. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, H.; Wang, J. Circular RNA CircSHKBP1 accelerates the proliferation, invasion, angiogenesis, and stem cell-like properties via modulation of microR-766-5p/high mobility group AT-hook 2 axis in laryngeal squamous cell carcinoma. Bioengineered 2022, 13, 11551–11563. [Google Scholar] [CrossRef]
- Yin, X.; Wang, J.; Shan, C.; Jia, Q.; Bian, Y.; Zhang, H. Circular RNA ZNF609 promotes laryngeal squamous cell carcinoma progression by upregulating epidermal growth factor receptor via sponging microRNA-134-5p. Bioengineered 2022, 13, 6929–6941. [Google Scholar] [CrossRef] [PubMed]
- Popov, T.M.; Stancheva, G.; Kyurkchiyan, S.G.; Petkova, V.; Panova, S.; Kaneva, R.P.; Popova, D.P. Global microRNA expression profile in laryngeal carcinoma unveils new prognostic biomarkers and novel insights into field cancerization. Sci. Rep. 2022, 12, 17051. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Lao, Z.; Zhang, H.; Wang, J.; Wang, S. Knockdown of circ_0001883 may inhibit epithelial-mesenchymal transition in laryngeal squamous cell carci-noma via the miR-125-5p/PI3K/AKT axis. Exp. Ther. Med. 2021, 22, 1007. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, C.; Zhou, P.; Chen, S.; Zheng, H. LncRNA CASC15 upregulates cyclin D1 by downregulating miR-365 in laryngeal squamous cell carcinoma to promote cell proliferation. J. Otolaryngol. Head Neck Surg. 2022, 51, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hu, X.; Huang, X. Long intergenic non-protein coding RNA 847 promotes laryngeal squamous cell carcinoma progression through the microRNA-181a-5p/zinc finger E-box binding homeobox 2 axis. Bioengineered 2022, 13, 9987–10000. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Ge, P. Long non-coding RNA HCG18 facilitates the progression of laryngeal and hypopharyngeal squamous cell carcinoma by upregulating FGFR1 via miR-133b. Mol. Med. Rep. 2021, 25, 46. [Google Scholar] [CrossRef]
- Shen, N.; Duan, X.; Feng, Y.; Zhang, J.; Qiao, X.; Ding, W. Long non-coding RNA HOXA11 antisense RNA upregulates spermatogenesis-associated serine-rich 2-like to enhance cisplatin resistance in laryngeal squamous cell carcinoma by suppressing microRNA-518a. Bioengineered 2022, 13, 974–984. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, P.; Cui, Y. Long noncoding RNA NEAT1 inhibits the acetylation of PTEN through the miR-524-5p /HDAC1 axis to promote the proliferation and invasion of laryngeal cancer cells. Aging 2021, 13, 24850–24865. [Google Scholar] [CrossRef]
- Han, L.; Zheng, C.; Wu, S. Long non-coding RNA NEAT1 promotes the malignancy of laryngeal squamous cell carcinoma by regulating the microRNA-204-5p/SEMA4B axis. Oncol. Lett. 2021, 22, 802. [Google Scholar] [CrossRef]
- Piotrowski, I.; Zhu, X.; Saccon, T.D.; Ashiqueali, S.; Schneider, A.; de Carvalho Nunes, A.D.; Noureddine, S.; Sobecka, A.; Barczak, W.; Szewczyk, M.; et al. miRNAs as Biomarkers for Diagnosing and Predicting Survival of Head and Neck Squamous Cell Car-cinoma Patients. Cancers 2021, 13, 3980. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Dai, L.; Wu, H.; Chen, C.; Ni, J.; Jin, E.; Zhou, X. Paired Box 5-Induced LINC00467 Upregulation Promotes the Progression of Laryngeal Squamous Cell Cancer by Triggering the MicroRNA-4735-3p/TNF Alpha-Induced Protein 3 Pathway. Mol. Biotechnol. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, B.; Lyu, K.; Yue, H.; Wei, F.; Xu, Y.; Chen, S.; Lin, Y.; Cai, Z.; Guo, X.; et al. Low expression of lncRNA SBF2-AS1 regulates the miR-302b-3p/TGFBR2 axis, promoting metastasis in laryngeal cancer. Mol. Carcinog. 2022, 61, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wan, L.; Gong, H.; Chen, S.; Kong, Y.; Xiao, B. Sevoflurane promotes the apoptosis of laryngeal squamous cell carcinoma in-vitro and inhibits its malignant progression via miR-26a/FOXO1 axis. Bioengineered 2021, 12, 6364–6376. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhang, Y.; Sun, H. Mechanism of miR-340-5p in laryngeal cancer cell proliferation and invasion through the lncRNA NEAT1/MMP11 axis. Pathol. Res. Pract. 2022, 236, 153912. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Li, J.; Ke, S. CircRNA CORO1C Regulates miR-654-3p/USP7 Axis to Mediate Laryngeal Squamous Cell Carcinoma Pro-gression. Biochem. Genet. 2022, 60, 1615–1629. [Google Scholar] [CrossRef]
- Fan, D.; Zhu, Y. Circ_0120175 promotes laryngeal squamous cell carcinoma development through up-regulating SLC7A11 by sponging miR-330-3p. Histochem. J. 2022, 53, 159–171. [Google Scholar] [CrossRef]
- Gao, S.; Xu, Q.; Zhou, Y.; Yi, Q. Serum levels of microRNA-21 and microRNA-10a can predict long-term prognosis in laryngeal cancer patients: A multicenter study. Transl. Cancer Res. 2020, 9, 3680–3690. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhu, H.; Gao, W. MicroRNA-330-3p represses the proliferation and invasion of laryngeal squamous cell car-cinoma through downregulation of Tra2beta-mediated Akt signaling. Mol. Cell. Probes 2020, 52, 101574. [Google Scholar] [CrossRef]
- Kyurkchiyan, S.G.; Popov, T.M.; Stancheva, G.; Rangachev, J.; Mitev, V.I.; Popova, D.P.; Kaneva, R.P. Novel insights into laryngeal squamous cell carcinoma from association study of aberrantly ex-pressed miRNAs, lncRNAs and clinical features in Bulgarian patients. J. BUON 2020, 25, 357–366. [Google Scholar]
- Gu, J.; Han, T.; Sun, L.; Yan, A.-H.; Jiang, X.-J. miR-552 promotes laryngocarcinoma cells proliferation and metastasis by targeting p53 pathway. Cell Cycle 2020, 19, 1012–1021. [Google Scholar] [CrossRef]
- Huang, Q.; Hsueh, C.; Guo, Y.; Wu, X.; Li, J.; Zhou, L. Lack of miR-1246 in small extracellular vesicle blunts tumorigenesis of laryngeal carcinoma cells by regulating Cyclin G2. IUBMB Life 2020, 72, 1491–1503. [Google Scholar] [CrossRef]
- Lin, X.-J.; Liu, H.; Li, P.; Wang, H.-F.; Yang, A.-K.; Di, J.-M.; Jiang, Q.-W.; Yang, Y.; Huang, J.-R.; Yuan, M.-L.; et al. miR-936 Suppresses Cell Proliferation, Invasion, and Drug Resistance of Laryngeal Squamous Cell Carcinoma and Targets GPR78. Front. Oncol. 2020, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-C.; Song, L.-Q.; Xu, W.-W.; Qi, J.-J.; Wang, X.-Y.; Su, Y. Serum miR-632 is a potential marker for the diagnosis and prognosis in laryngeal squamous cell carcinoma. Acta Oto-Laryngologica 2020, 140, 418–421. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, C.; Zhang, A.; Lu, C.; Liu, L. Overexpression of circRNA_100290 promotes the progression of laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C axis. Biomed. Pharmacother. 2020, 125, 109874. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qian, J.; Xia, X.; Ye, B. Long non-coding RNA OIP5-AS1 serves as an oncogene in laryngeal squamous cell carcinoma by regulating miR-204-5p/ZEB1 axis. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 2177–2184. [Google Scholar] [CrossRef]
- Wang, J.-X.; Jia, X.-J.; Liu, Y.; Dong, J.-H.; Ren, X.-M.; Xu, O.; Liu, S.-H.; Shan, C.-G. Silencing of miR-17-5p suppresses cell proliferation and promotes cell apoptosis by directly targeting PIK3R1 in laryngeal squamous cell carcinoma. Cancer Cell Int. 2020, 20, 14. [Google Scholar] [CrossRef] [Green Version]
- Xun, W.; Cen, W.; Dahai, Y.; Huaqing, W.; Jiping, S.; Mengzhu, G.; Ning, M. LncRNA miR143HG suppresses miR-21 through methylation to inhibit cell invasion and migration. Laryngoscope 2020, 130, E640–E645. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Q.; Li, F. miR-140-5p targeted FGF9 and inhibited the cell growth of laryngeal squamous cell carcinoma. Biochem. Cell Biol. 2020, 98, 83–89. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Wang, L.; Xu, P.-C.; Huang, F.-J.; Jian, X.; Wei, Z.-C.; Chen, Y.-Q. LINC01605 promotes the proliferation of laryngeal squamous cell carcinoma through targeting miR-493-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10379–10386. [Google Scholar] [PubMed]
- Li, P.; Lin, X.-J.; Yang, Y.; Yang, A.-K.; Di, J.-M.; Jiang, Q.-W.; Huang, J.-R.; Yuan, M.-L.; Xing, Z.-H.; Wei, M.-N.; et al. Reciprocal regulation of miR-1205 and E2F1 modulates progression of laryngeal squamous cell carcinoma. Cell Death Dis. 2019, 10, 916. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.; Li, L.; Wang, C.; Zhang, Q.; He, S. MiR-381-3p suppresses biological characteristics of cancer in head-neck squamous cell carcinoma cells by tar-geting nuclear autoantigenic sperm protein (NASP). Biosci. Biotechnol. Biochem. 2020, 84, 703–713. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Z.; Zhang, K.; Dong, Y.; Zhang, A.; Lu, C.; Liu, L. MicroRNA-107 inhibits proliferation and invasion of laryngeal squamous cell carcinoma cells by targeting CACNA2D1 in vitro. Anti-Cancer Drugs 2020, 31, 260–271. [Google Scholar] [CrossRef]
- Chen, L.; Sun, D.Z.; Fu, Y.G.; Yang, P.Z.; Lv, H.Q.; Gao, Y.; Zhang, X.Y. Upregulation of microRNA-141 suppresses epithelial-mesenchymal transition and lymph node metastasis in laryngeal cancer through HOXC6-dependent TGF-beta signaling pathway. Cell Signal 2020, 66, 109444. [Google Scholar] [CrossRef]
- Fang, R.; Huang, Y.; Xie, J.; Zhang, J.; Ji, X. Downregulation of miR-29c-3p is associated with a poor prognosis in patients with laryngeal squamous cell carcinoma. Diagn. Pathol. 2019, 14, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, X.; Shen, N.; Chen, J.; Wang, J.; Zhu, Q.; Zhai, Z. Circular RNA MYLK serves as an oncogene to promote cancer progression via microRNA-195/cyclin D1 axis in laryngeal squamous cell carcinoma. Biosci. Rep. 2019, 39, BSR20190227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Xi, K. LncRNA RGMB-AS1 promotes laryngeal squamous cell carcinoma cells progression via sponging miR-22/NLRP3 axis. Biomed. Pharmacother. 2019, 118, 109222. [Google Scholar] [CrossRef]
- Zhu, M.; Zhang, C.; Chen, D.; Chen, S.; Zheng, H. MicroRNA-98-HMGA2-POSTN signal pathway reverses epithelial-to-mesenchymal transition in laryngeal squamous cell carcinoma. Biomed. Pharmacother. 2019, 117, 108998. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.T.; Tong, X.; Zhang, Z.X.; Sun, Y.Y.; Yan, W.; Xu, Z.M.; Fu, W.N. MYCT1 represses apoptosis of laryngeal cancerous cells through the MAX/miR-181a/NPM1 pathway. FEBS J. 2019, 286, 3892–3908. [Google Scholar] [CrossRef]
- Grzelczyk, W.L.; Szemraj, J.; Kwiatkowska, S.; Józefowicz-Korczyńska, M. Serum expression of selected miRNAs in patients with laryngeal squamous cell carcinoma (LSCC). Diagn. Pathol. 2019, 14, 49. [Google Scholar] [CrossRef]
- Yuan, Z.; Xiu, C.; Liu, D.; Zhou, G.; Yang, H.; Pei, R. Long noncoding RNA LINC-PINT regulates laryngeal carcinoma cell stemness and chemoresistance through miR-425-5p/PTCH1/SHH axis. J. Cell Physiol. 2019, 234, 23111–23122. [Google Scholar] [CrossRef]
- Chen, X.; Su, X.; Zhu, C.; Zhou, J. Knockdown of hsa_circ_0023028 inhibits cell proliferation, migration, and invasion in laryngeal cancer by sponging miR-194-5p. Biosci. Rep. 2019, 39, BSR20190177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapa RM, L.; Barros-Filho, M.C.; Marchi, F.A.; Domingues MA, C.; de Carvalho, G.B.; Drigo, S.A. Integrated miRNA and mRNA expression analysis uncovers drug targets in laryngeal squamous cell car-cinoma patients. Oral Oncol. 2019, 93, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, J.; Guo, Y.-N.; Yang, B.-B. LncRNA SNHG20 promotes the development of laryngeal squamous cell carcinoma by regulating miR-140. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3401–3409. [Google Scholar] [PubMed]
- Cui, W.; Meng, W.; Zhao, L.; Cao, H.; Chi, W.; Wang, B. TGF-beta-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int. J. Oncol. 2019, 54, 2005–2018. [Google Scholar]
- Xiao, D.; Cui, X.; Wang, X. Long noncoding RNA XIST increases the aggressiveness of laryngeal squamous cell carcinoma by regulating miR-124-3p/EZH2. Exp. Cell Res. 2019, 381, 172–178. [Google Scholar] [CrossRef]
- Tian, L.; Cao, J.; Jiao, H.; Zhang, J.; Ren, X.; Liu, X.; Liu, M.; Sun, Y. CircRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin. Sci. 2019, 133, 1053–1066. [Google Scholar] [CrossRef]
- Gao, C.; Hu, S. miR-506 is a YAP1-dependent tumor suppressor in laryngeal squamous cell carcinoma. Cancer Biol. Ther. 2019, 20, 826–836. [Google Scholar] [CrossRef]
- Li, Y.; Tao, C.; Dai, L.; Cui, C.; Chen, C.; Wu, H.; Wei, Q.; Zhou, X. MicroRNA-625 inhibits cell invasion and epithelial–mesenchymal transition by targeting SOX4 in laryngeal squamous cell carcinoma. Biosci. Rep. 2019, 39, BSR20181882. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Sun, J.; Cao, H. MicroRNA-384 regulates cell proliferation and apoptosis through directly targeting WISP1 in laryngeal cancer. J. Cell. Biochem. 2019, 120, 3018–3026. [Google Scholar] [CrossRef]
- Zhang, F.; Cao, H. MicroRNA-143-3p suppresses cell growth and invasion in laryngeal squamous cell carcinoma via tar-geting the k-Ras/Raf/MEK/ERK signaling pathway. Int. J. Oncol. 2019, 54, 689–701. [Google Scholar]
- Luo, M.; Sun, G.; Sun, J.-W. MiR-196b affects the progression and prognosis of human LSCC through targeting PCDH-17. Auris Nasus Larynx 2019, 46, 583–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Zhang, C.; Li, W.; Li, H.; Sang, J.; Zhao, Q.; Bo, Y.; Luo, H.; Zheng, X.; Lu, Y.; et al. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Mol. Ther. 2019, 27, 365–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Tang, M.; Xu, X.; Jiang, B.; Wei, Y.; Qian, H.; Lu, X. MiR-143-3p suppresses cell proliferation, migration, and invasion by targeting Melanoma-Associated Antigen A9 in laryngeal squamous cell carcinoma. J. Cell. Biochem. 2018, 120, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Xia, X.; Zhu, Y.; Diao, W.; Zhu, X.; Gao, Z.; Chen, X. Circular RNA Expression Profile in Laryngeal Squamous Cell Carcinoma Revealed by Microarray. Cell. Physiol. Biochem. 2018, 50, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, H.; Zhao, Y.; Zhao, Y. CDR1as is overexpressed in laryngeal squamous cell carcinoma to promote the tumour’s progression via miR-7 signals. Cell Prolif. 2018, 51, e12521. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Zhang, D.; Huang, H.; Yu, Y.; Yang, Z.; Niu, Y.; Liu, J. MicroRNA-1225-5p acts as a tumor-suppressor in laryngeal cancer via targeting CDC14B. Biol. Chem. 2019, 400, 237–246. [Google Scholar] [CrossRef]
- Zhao, R.; Li, F.Q.; Tian, L.L.; Shang, D.S.; Guo, Y.; Zhang, J.R.; Liu, M. Comprehensive analysis of the whole coding and non-coding RNA transcriptome expression profiles and construction of the circRNA-lncRNA co-regulated ceRNA network in laryngeal squamous cell carcinoma. Funct. Integr. Genom. 2019, 19, 109–121. [Google Scholar] [CrossRef]
- Feng, W.-T.; Yao, R.; Xu, L.-J.; Zhong, X.-M.; Liu, H.; Sun, Y.; Zhou, L.-L. Effect of miR-363 on the proliferation, invasion and apoptosis of laryngeal cancer by targeting Mcl-1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4564–4572. [Google Scholar]
- Zhao, X.; Zhang, W.; Ji, W. MYO5A inhibition by miR-145 acts as a predictive marker of occult neck lymph node metastasis in human laryngeal squamous cell carcinoma. OncoTargets Ther. 2018, ume 11, 3619–3635. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.-T.; Zhang, L.-J.; Huang, Y.-W.; Li, C.; Jiang, N.; Niu, Y.-J. MiR-154 inhibits the growth of laryngeal squamous cell carcinoma by targeting GALNT7. Biochem. Cell Biol. 2018, 96, 752–760. [Google Scholar] [CrossRef]
- Hui, L.; Zhang, J.; Guo, X. MiR-125b-5p suppressed the glycolysis of laryngeal squamous cell carcinoma by down-regulating hexokinase-2. Biomed. Pharmacother. 2018, 103, 1194–1201. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Tang, S. MicroRNA-4497 functions as a tumor suppressor in laryngeal squamous cell carcinoma via negatively modulation the GBX2. Auris Nasus Larynx 2019, 46, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, W.; Ji, W. miR-196b is a prognostic factor of human laryngeal squamous cell carcinoma and promotes tumor progression by targeting SOCS2. Biochem. Biophys. Res. Commun. 2018, 501, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, J.; Ge, W.; Jiang, Y. Long non-coding RNA LOC554202 promotes laryngeal squamous cell carcinoma progression through regu-lating miR-31. J. Cell. Biochem. 2018, 119, 6953–6960. [Google Scholar] [CrossRef]
- Zhou, Z.-X.; Zhang, Z.-P.; Tao, Z.-Z.; Tan, T.-Z. miR-632 Promotes Laryngeal Carcinoma Cell Proliferation, Migration, and Invasion Through Negative Regulation of GSK3β. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2020, 28, 21–31. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, W.; Ji, W. YB-1 promotes laryngeal squamous cell carcinoma progression by inducing miR-155 expression via c-Myb. Futur. Oncol. 2018, 14, 1579–1589. [Google Scholar] [CrossRef]
- Pang, H.; Xu, X.; Dai, L.; Wang, K.; Yao, X. MicroRNA-195 is associated with regulating the pathophysiologic process of human laryngeal squamous cell carcinoma. Mol. Med. Rep. 2018, 17, 5283–5291. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Lu, B.; Li, X.; Miao, W.; Li, J.; Shi, Y.; Yu, W. MicroRNA-26a inhibits proliferation and tumorigenesis via targeting CKS2 in laryngeal squamous cell carcinoma. Clin. Exp. Pharmacol. Physiol. 2018, 45, 444–451. [Google Scholar] [CrossRef]
- Wang, J.; Yang, S.; Ge, W.; Wang, Y.; Han, C.; Li, M. MiR-613 suppressed the laryngeal squamous cell carcinoma progression through regulating PDK1. J. Cell. Biochem. 2018, 119, 5118–5125. [Google Scholar] [CrossRef]
- Re, M.; Magliulo, G.; Gioacchini, F.M.; Bajraktari, A.; Bertini, A.; Çeka, A.; Rubini, C.; Ferrante, L.; Procopio, A.D.; Olivieri, F. Expression Levels and Clinical Significance of miR-21-5p, miR-let-7a, and miR-34c-5p in Laryngeal Squamous Cell Carcinoma. BioMed. Res. Int. 2017, 2017, 3921258. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, J.; Wang, L.; Yang, X.; Liu, X. MicroRNA-195 inhibits cell proliferation, migration and invasion in laryngeal squamous cell carcinoma by tar-geting ROCK1. Mol. Med. Rep. 2017, 16, 7154–7162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuang, Y.; Zhou, X.; Li, C.; Huang, Y.; Zhang, L. MicroRNA-503 serves an oncogenic role in laryngeal squamous cell carcinoma via targeting programmed cell death protein 4. Mol. Med. Rep. 2017, 16, 5249–5256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Wu, Y.; He, X.; Zhang, C.; Zhu, M.; Chen, B.; Liu, Q.; Binquan, W.; Xiaoling, H.; Wen, S.; et al. MicroRNA-204-5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box C1. J. Cancer 2017, 8, 2356–2368. [Google Scholar] [CrossRef]
- Shuang, Y.; Li, C.; Zhou, X.; Huang, Y.; Zhang, L. MicroRNA-195 inhibits growth and invasion of laryngeal carcinoma cells by directly targeting DCUN1D1. Oncol. Rep. 2017, 38, 2155–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuang, Y.; Li, C.; Zhou, X.; Huang, Y.-W.; Zhang, L. Expression of miR-195 in laryngeal squamous cell carcinoma and its effect on proliferation and apoptosis of Hep-2. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3232–3238. [Google Scholar] [PubMed]
- Wang, S.; Guo, D.; Li, C. Downregulation of miRNA-26b inhibits cancer proliferation of laryngeal carcinoma through autophagy by targeting ULK2 and inactivation of the PTEN/AKT pathway. Oncol. Rep. 2017, 38, 1679–1687. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Fan, Y.; Ayiheng, Q.; Zhang, H.; Yong, J.; Hu, B. MicroRNA-125b targeted STAT3 to inhibit laryngeal squamous cell carcinoma cell growth and motility. Oncol. Lett. 2017, 14, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hu, H. Long non-coding RNA CCAT1/miR-218/ZFX axis modulates the progression of laryngeal squamous cell cancer. Tumour. Biol. 2017, 39, 1010428317699417. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Lu, E.; Lu, L.; Zhang, C. MiR-29a-3p suppresses cell proliferation in laryngocarcinoma by targeting prominin 1. FEBS Open Bio 2017, 7, 645–651. [Google Scholar] [CrossRef]
- Hussein, S.; Mosaad, H.; Rashed, H.E.; El-Anwar, M.W. Up-regulated miR-221 expression as a molecular diagnostic marker in laryngeal squamous cell carcinoma and its correlation with Apaf-1 expression. Cancer Biomarkers 2017, 19, 279–287. [Google Scholar] [CrossRef]
- Bruzgielewicz, A.; Osuch-Wojcikiewicz, E.; Niemczyk, K.; Sieniawska-Buccella, O.; Siwak, M.; Walczak, A.; Nowak, A.; Majsterek, I. Altered Expression of miRNAs Is Related to Larynx Cancer TNM Stage and Patients’ Smoking Status. DNA Cell Biol. 2017, 36, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Long, X.-B.; Sun, G.-B.; Hu, S.; Liang, G.-T.; Wang, N.; Zhang, X.-H.; Cao, P.-P.; Zhen, H.-T.; Cui, Y.-H. Let-7a microRNA functions as a potential tumor suppressor in human laryngeal cancer. Oncol. Rep. 2009, 22, 1189–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Zhu, D.; Liu, M.; Sun, Y.; Tian, L. Downregulation of miR-21 modulates Ras expression to promote apoptosis and suppress invasion of Laryngeal squamous cell carcinoma. Eur. J. Cancer 2010, 46, 3409–3416. [Google Scholar] [CrossRef]
- Shen, Z.; Zhan, G.; Ye, D.; Ren, Y.; Cheng, L.; Wu, Z.; Guo, J. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med. Oncol. 2012, 29, 2473–2480. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Fu, W.; Chen, H.; Shang, C.; Zhong, M. miR-24 functions as a tumor suppressor in Hep2 laryngeal carcinoma cells partly through down-regulation of the S100A8 protein. Oncol. Rep. 2011, 27, 1097–1103. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, M.; Wang, C.; Lin, C.; Sun, Y.; Jin, D. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer. Res. 2011, 31, 3859–3863. [Google Scholar]
- Cai, K.; Wang, Y.; Bao, X. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J. Exp. Clin. Cancer Res. 2011, 30, 73. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.D.; Zhang, W.; Liang, H.J.; Ji, W.Y. Overexpression of miR -155 promotes proliferation and invasion of human laryngeal squamous cell carci-noma via targeting SOCS1 and STAT3. PLoS ONE 2013, 8, e56395. [Google Scholar]
- Li, X.; Wang, H.-L.; Peng, X.; Zhou, H.-F.; Wang, X. miR-1297 mediates PTEN expression and contributes to cell progression in LSCC. Biochem. Biophys. Res. Commun. 2012, 427, 254–260. [Google Scholar] [CrossRef]
- Cao, P.; Zhou, L.; Zhang, J.; Zheng, F.; Wang, H.; Ma, D.; Tian, J. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck 2013, 35, 720–728. [Google Scholar] [CrossRef]
- Saito, K.; Inagaki, K.; Kamimoto, T.; Ito, Y.; Sugita, T.; Nakajo, S.; Hirasawa, A.; Iwamaru, A.; Ishikura, T.; Hanaoka, H.; et al. MicroRNA-196a Is a Putative Diagnostic Biomarker and Therapeutic Target for Laryngeal Cancer. PLoS ONE 2013, 8, e71480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, X.; Kai, W.; Wei, G.; Chunming, Z.; Fuhui, H.; Shuxin, W.; Binquan, W. MicroRNA-106b regulates the tumor suppressor RUNX3 in laryngeal carcinoma cells. FEBS Lett. 2013, 587, 3166–3174. [Google Scholar] [CrossRef]
- Ayaz, L.; Görür, A.; Yaroğlu, H.Y.; Özcan, C.; Tamer, L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: Potential early-detection markers for laryngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2013, 139, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.-N.; Wang, Z.-H.; Sheng, Y.; Zhang, Q.; Yan, J.; Hou, J.; Zhu, K.; Xu, Y.-L.; Zhang, X.-H.; Xu, M.; et al. miR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med. Oncol. 2014, 31, 789. [Google Scholar] [CrossRef]
- Shen, Z.; Zhan, G.; Deng, H.; Ren, Y.; Ye, D.; Xiao, B.; Guo, J. MicroRNA-519a demonstrates significant tumour suppressive activity in laryngeal squamous cells by targeting anti-carcinoma HuR gene. J. Laryngol. Otol. 2013, 127, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Zhang, J.; Ge, J.; Xiao, H.; Lu, J.; Fu, S.; Liu, M.; Sun, Y. MicroRNA-205 suppresses proliferation and promotes apoptosis in laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 785. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, M.; Tao, Z.; Hua, Q.; Chen, S.; Xiao, B. Identification of predictive biomarkers for early diagnosis of larynx carcinoma based on microRNA expression data. Cancer Genet. 2013, 206, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tian, L.; Wang, L.; Yao, H.; Zhang, J.; Lu, J.; Sun, Y.; Gao, X.; Xiao, H.; Liu, M. Down-Regulation of miR-129-5p Inhibits Growth and Induces Apoptosis in Laryngeal Squamous Cell Carcinoma by Targeting APC. PLoS ONE 2013, 8, e77829. [Google Scholar] [CrossRef] [Green Version]
- Yungang, W.; Xiaoyu, L.; Pang, T.; Wenming, L.; Pan, X. miR-370 targeted FoxM1 functions as a tumor suppressor in laryngeal squamous cell carcinoma (LSCC). Biomed. Pharmacother. 2014, 68, 149–154. [Google Scholar] [CrossRef]
- Zhang, T.; Han, G.; Wang, Y.; Chen, K.; Sun, Y. MicroRNA expression profiles in supraglottic carcinoma. Oncol. Rep. 2014, 31, 2029–2034. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wang, Z.-M.; Song, Y.; Tai, X.-H.; Ji, W.-Y.; Gu, H. MicroRNA-126 modulates the tumor microenvironment by targeting calmodulin-regulated spectrin-associated protein 1 (Camsap1). Int. J. Oncol. 2014, 44, 1678–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.-Y.; Zhang, T.-H.; Qu, L.-M.; Feng, J.-P.; Tian, L.-L.; Zhang, B.-H.; Li, D.-D.; Sun, Y.-N.; Liu, M. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int. J. Clin. Exp. Pathol. 2014, 7, 56–63. [Google Scholar]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 148. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Tian, Y.; Bai, W.-L.; Ma, X.-L. Expression and clinical significance of microRNA-152 in supragalottic laryngeal carcinoma. Tumor Biol. 2014, 35, 11075–11079. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wu, Y.; Liu, Y.; Deng, H.; Shen, Z.; Xiao, B.; Guo, J. miR-21, miR-106b and miR-375 as Novel Potential Biomarkers for Laryngeal Squamous Cell Carcinoma. Curr. Pharm. Biotechnol. 2014, 15, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.-M.; Lin, Y.-F.; Jiang, L.; Chen, L.-S.; Luo, X.-N.; Song, X.-H.; Chen, S.-H.; Zhang, S.-Y. Micro-ribonucleic acid expression profiling and bioinformatic target gene analyses in laryngeal carcinoma. OncoTargets Ther. 2014, 7, 525–533. [Google Scholar] [CrossRef] [Green Version]
- Hu, A.; Huang, J.-J.; Xu, W.-H.; Jin, X.-J.; Li, J.-P.; Tang, Y.-J.; Huang, X.-F.; Cui, H.-J.; Sun, G.-B. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: Association with patient survival. Am. J. Transl. Res. 2014, 6, 604–613. [Google Scholar]
- Xiao, X.; Zhou, L.; Cao, P.; Gong, H.; Zhang, Y. MicroRNA-93 regulates cyclin G2 expression and plays an oncogenic role in laryngeal squamous cell carcinoma. Int. J. Oncol. 2015, 46, 161–174. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Wu, J.; Li, Z.; Qin, H.; Wang, B.; Wong, T.S.; Yang, W.; Fu, Q.-L.; Lei, W. miR-375 Suppresses IGF1R Expression and Contributes to Inhibition of Cell Progression in Laryngeal Squamous Cell Carcinoma. BioMed. Res. Int. 2014, 2014, 374598. [Google Scholar] [CrossRef]
- Wu, S.; Jia, S.; Xu, P. MicroRNA-9 as a novel prognostic biomarker in human laryngeal squamous cell carcinoma. Int. J. Clin. Exp. Med. 2014, 7, 5523–5528. [Google Scholar]
- Re, M.; Ceka, A.; Rubini, C.; Ferrante, L.; Zizzi, A.; Gioacchini, F.M.; Tulli, M.; Spazzafumo, L.; Sellari-Franceschini, S.; Procopio, A.D.; et al. MicroRNA-34c-5p is related to recurrence in laryngeal squamous cell carcinoma. Laryngoscope 2015, 125, E306–E312. [Google Scholar] [CrossRef]
- Xu, L.; Chen, Z.; Xue, F.; Chen, W.; Ma, R.; Cheng, S.; Cui, P. MicroRNA-24 inhibits growth, induces apoptosis, and reverses radioresistance in laryngeal squamous cell car-cinoma by targeting X-linked inhibitor of apoptosis protein. Cancer Cell Int. 2015, 15, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karatas, O.F.; Yuceturk, B.; Suer, I.; Yilmaz, M.; Cansiz, H.; Solak, M.; Ittmann, M.; Ozen, M. Role of miR-145 in human laryngeal squamous cell carcinoma. Head Neck 2016, 38, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.S.; Guzel, E.; Karatas, O.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. MiR-221 as a pre- and postoperative plasma biomarker for larynx cancer patients. Laryngoscope 2015, 125, E377–E381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-W.; Liu, N.; Chen, S.; Wang, Y.; Zhang, Z.-X.; Sun, Y.-Y.; Qiu, G.-B.; Fu, W.-N. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn. Pathol. 2015, 10, 22. [Google Scholar] [CrossRef] [Green Version]
- Roncon, P.; Soukupovà, M.; Binaschi, A.; Falcicchia, C.; Zucchini, S.; Ferracin, M.; Langley, S.R.; Petretto, E.; Johnson, M.R.; Marucci, G.; et al. MicroRNA profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy—Comparison with human epileptic samples. Sci. Rep. 2015, 5, 14143. [Google Scholar] [CrossRef] [Green Version]
- Maia, D.; De Carvalho, A.C.; Horst, M.A.; Carvalho, A.L.; Scapulatempo-Neto, C.; Vettore, A.L. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J. Transl. Med. 2015, 13, 262. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.F.; Wang, H.M.; Lu, B.C.; Zhang, G.Z.; Ma, H.M.; Wu, Z.Y. miR-206 inhibits human laryngeal squamous cell carcinoma cell growth by regulation of cyclinD2. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 2697–2702. [Google Scholar]
- Hu, A.; Huang, J.J.; Xu, W.H.; Jin, X.J.; Li, J.P.; Tang, Y.J. MiR-21/miR-375 ratio is an independent prognostic factor in patients with laryngeal squamous cell carcinoma. Am. J. Cancer Res. 2015, 5, 1775–1785. [Google Scholar]
- Zhang, X.-W.; Liu, N.; Chen, S.; Wang, Y.; Sun, K.-L.; Xu, Z.-M.; Fu, W.-N. Upregulation of microRNA-23a regulates proliferation and apoptosis by targeting APAF-1 in laryngeal carcinoma. Oncol. Lett. 2015, 10, 410–416. [Google Scholar] [CrossRef] [Green Version]
- Janiszewska, J.; Szaumkessel, M.; Kostrzewska-Poczekaj, M.; Bednarek, K.; Paczkowska, J.; Jackowska, J. Global miRNA Expression Profiling Identifies miR-1290 as Novel Potential oncomiR in Laryngeal Car-cinoma. PLoS ONE 2015, 10, e0144924. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, G.; Lu, B.; Li, J.; Wu, Z.; Ma, H.; Wang, H.; Lian, R. miR-340 impedes the progression of laryngeal squamous cell carcinoma by targeting EZH2. Gene 2016, 577, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Gao, W.; Zhang, C.; Wen, S.; Huangfu, H.; Kang, J.; Wang, B. Hsa-miR-301a-3p Acts as an Oncogene in Laryngeal Squamous Cell Carcinoma via Target Regulation of Smad4. J. Cancer 2015, 6, 1260–1275. [Google Scholar] [CrossRef] [Green Version]
- Zhong, G.; Xiong, X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Biol. Res. 2015, 48, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marioni, G.; Agostini, M.; Cappellesso, R.; Bedin, C.; Ottaviano, G.; Marchese-Ragona, R.; Lovato, A.; Cacco, T.; Giacomelli, L.; Nitti, D.; et al. miR-19a and SOCS-1 expression in the differential diagnosis of laryngeal (glottic) verrucous squamous cell carcinoma. J. Clin. Pathol. 2016, 69, 415–421. [Google Scholar] [CrossRef]
- Gao, S.; Wang, J.; Xie, J.; Zhang, T.; Dong, P. Role of miR-138 in the regulation of larynx carcinoma cell metastases. Tumor Biol. 2015, 37, 15601–15606. [Google Scholar] [CrossRef]
- Karatas, O.F.; Suer, I.; Yuceturk, B.; Yilmaz, M.; Hajiyev, Y.; Creighton, C.J.; Ittmann, M.; Ozen, M. The role of miR-145 in stem cell characteristics of human laryngeal squamous cell carcinoma Hep-2 cells. Tumor Biol. 2016, 37, 4183–4192. [Google Scholar] [CrossRef]
- Cybula, M.; Wieteska, L.; Józefowicz-Korczyńska, M.; Karbownik, M.S.; Grzelczyk, W.L.; Szemraj, J. New miRNA expression abnormalities in laryngeal squamous cell carcinoma. Cancer Biomarkers 2016, 16, 559–568. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-L.; Wang, X.; Yang, D.; Shi, W.-J. The Expression of MicroRNA-155 in Plasma and Tissue Is Matched in Human Laryngeal Squamous Cell Carcinoma. Yonsei Med. J. 2016, 57, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-Y.; Lu, J.-B.; Xu, Y. MicroRNA-153 inhibits the proliferation and invasion of human laryngeal squamous cell carcinoma by targeting KLF5. Exp. Ther. Med. 2016, 11, 2503–2508. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; An, R.; Zhao, R.; Sun, Y.; Liu, M.; Tian, L. miR-375 exhibits a more effective tumor-suppressor function in laryngeal squamous carcinoma cells by regulating KLF4 expression compared with simple co-transfection of miR-375 and miR-206. Oncol. Rep. 2016, 36, 952–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Li, L.; Feng, P.; Wan, J.; Li, J. Downregulation of miR-34a contributes to the proliferation and migration of laryngeal carcinoma cells by targeting cyclin D1. Oncol. Rep. 2016, 36, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cui, C.-L.; Chen, W.-L.; Fu, Z.-Y.; Cui, X.-Y.; Gong, X. miR-144 suppresses the growth and metastasis of laryngeal squamous cell carcinoma by targeting IRS1. Am. J. Transl. Res. 2016, 8, 1–11. [Google Scholar] [PubMed]
- Wei, L.; Mao, M.; Liu, H. Droplet digital PCR and qRT-PCR to detect circulating miR-21 in laryngeal squamous cell car-cinoma and pre-malignant laryngeal lesions. Acta Otolaryngol. 2016, 136, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Xu, L.; Chen, T.; Dong, P. MiR-203 is involved in the laryngeal carcinoma pathogenesis via targeting VEGFA and Cox-2. OncoTargets Ther. 2016, ume 9, 4629–4637. [Google Scholar] [CrossRef] [Green Version]
- Erkul, E.; Yilmaz, I.; Gungor, A.; Kurt, O.; Babayigit, M.A. MicroRNA-21 in laryngeal squamous cell carcinoma: Diagnostic and prognostic features. Laryngoscope 2017, 127, E62–E66. [Google Scholar] [CrossRef]
- Li, J.Z.-H.; Gao, W.; Lei, W.-B.; Zhao, J.; Chan, J.Y.-W.; Wei, W.I.; Ho, W.-K.; Wong, T.-S. MicroRNA 744-3p promotes MMP-9-mediated metastasis by simultaneously suppressing PDCD4 and PTEN in laryngeal squamous cell carcinoma. Oncotarget 2016, 7, 58218–58233. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Yu, J.; Ma, Y.; Wang, F.; Liu, H. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol. Lett. 2016, 12, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Lin, Y.-P.; Yang, D.; Zhang, G.; Zhou, H.-F. Clinical Significance of miR-149 in the Survival of Patients with Laryngeal Squamous Cell Carcinoma. BioMed. Res. Int. 2016, 2016, 8561251. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Tang, Q.; Li, S.; Yang, X. Inhibition of HAX-1 by miR-125a reverses cisplatin resistance in laryngeal cancer stem cells. Oncotarget 2016, 7, 86446–86456. [Google Scholar] [CrossRef] [Green Version]
- Song, F.-C.; Yang, Y.; Liu, J.-X. Expression and significances of MiRNA Let-7 and HMGA2 in laryngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4452–4458. [Google Scholar]
- Jili, S.; Eryong, L.; Lijuan, L.; Chao, Z. RUNX3 inhibits laryngeal squamous cell carcinoma malignancy under the regulation of miR-148a-3p/DNMT1 axis. Cell Biochem. Funct. 2016, 34, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Lian, R.; Lu, B.; Jiao, L.; Li, S.; Wang, H.; Miao, W.; Yu, W. MiR-132 plays an oncogenic role in laryngeal squamous cell carcinoma by targeting FOXO1 and activating the PI3K/AKT pathway. Eur. J. Pharmacol. 2016, 792, 1–6. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Li, Y.-Z. MicroRNA-133a suppresses the proliferation, migration, and invasion of laryngeal carcinoma cells by targeting CD47. Tumor Biol. 2016, 37, 16103–16113. [Google Scholar] [CrossRef]
- Ge, W.; Han, C.; Wang, J.; Zhang, Y. MiR-300 suppresses laryngeal squamous cell carcinoma proliferation and metastasis by targeting ROS1. Am. J. Transl. Res. 2016, 8, 3903–3911. [Google Scholar] [PubMed]
- Lu, E.; Su, J.; Zeng, W.; Zhang, C. Enhanced miR-9 promotes laryngocarcinoma cell survival via down-regulating PTEN. Biomed. Pharmacother. 2016, 84, 608–613. [Google Scholar] [CrossRef]
- Chen, S.; Sun, Y.Y.; Zhang, Z.X.; Li, Y.H.; Xu, Z.M.; Fu, W.N. Transcriptional suppression of microRNA-27a contributes to laryngeal cancer differentiation via GSK-3beta-involved Wnt/beta-catenin pathway. Oncotarget 2017, 8, 14708–14718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Lv, K.; Chen, W.; Zhao, J.; Luo, J.; Wu, J. miR-375 and miR-205 Regulate the Invasion and Migration of Laryngeal Squamous Cell Carcinoma Syner-gistically via AKT-Mediated EMT. Biomed. Res. Int. 2016, 2016, 9652789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Lin, Y.-P.; Yang, D.; Zhang, G.; Zhou, H.-F. Expression of serum microRNA-378 and its clinical significance in laryngeal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5137–5142. [Google Scholar]
- He, F.-Y.; Liu, H.-J.; Guo, Q.; Sheng, J.-L. Reduced miR-300 expression predicts poor prognosis in patients with laryngeal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 760–764. [Google Scholar]
- Shuang, Y.; Yao, X.; Liu, J.; Niu, J.; Guo, W.; Li, C. Serum-derived extracellular vesicles mediate Smad4 expression through shuttling microRNA-27a in the progression of laryngeal squamous cell carcinoma. Hum. Cell 2022, 35, 1084–1099. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Gu, D.; Li, P. LncRNA SNHG16 promotes proliferation and migration in laryngeal squamous cell carcinoma via the miR-140-5p/NFAT5/Wnt/beta-catenin pathway axis. Pathol. Res. Pract. 2022, 229, 153727. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, Y.; Chen, X.; Fan, J.; Zheng, W.; Cao, C. miR-206 Inhibits Laryngeal Carcinoma Cell Multiplication, Migration, and Invasion. J. Healthc. Eng. 2021, 2021, 5614861. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Ren, G.; Xu, J.; Yin, C.; Shi, Y. LncRNA MNX1-AS1 Contributes to Laryngeal Squamous Cell Carcinoma Growth and Migration by Regulating mir-744-5p/bcl9/beta-Catenin Axis. Cell Transpl. 2021, 30, 9636897211005682. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yu, H.; Yu, T.; Xiao, D.; Wang, X. LncRNA MNX1-AS1 drives aggressive laryngeal squamous cell carcinoma progression and serves as a ceRNA to target FoxM1 by sponging microRNA-370. Aging 2021, 13, 9900–9910. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Song, Y.; Tai, X.; Liu, B.; Ji, W. MicroRNA expression and its detection in human supraglottic laryngeal squamous cell carcinoma. Biomed. Rep. 2013, 1, 743–746. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.; Qi, Z. Clinical significance of miRNA-195 expression in patients with laryngeal carcinoma. J. BUON 2019, 24, 315–322. [Google Scholar]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Lan, H.; Lu, H.; Wang, X.; Jin, H. MicroRNAs as Potential Biomarkers in Cancer: Opportunities and Challenges. BioMed. Res. Int. 2015, 2015, 125094. [Google Scholar] [CrossRef] [Green Version]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer—A brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dika, E.; Riefolo, M.; Porcellini, E.; Broseghini, E.; Ribero, S.; Senetta, R.; Osella-Abate, S.; Scarfì, F.; Lambertini, M.; Veronesi, G.; et al. Defining the Prognostic Role of MicroRNAs in Cutaneous Melanoma. J. Investig. Dermatol. 2020, 140, 2260–2267. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Nakajima, G.O.; Gavin, E.; Morris, C.G.; Kudo, K.; Hayashi, K.; Ju, J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraf-fin-embedded samples. RNA 2007, 13, 1668–1674. [Google Scholar] [CrossRef] [Green Version]
- Peskoe, S.B.; Barber, J.R.; Zheng, Q.; Meeker, A.K.; De Marzo, A.M.; Platz, E.A.; Lupold, S.E. Differential long-term stability of microRNAs and RNU6B snRNA in 12–20 year old archived formalin-fixed paraffin-embedded specimens. BMC Cancer 2017, 17, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Thera-peutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Gorphe, P. A comprehensive review of Hep-2 cell line in translational research for laryngeal cancer. Am. J. Cancer Res. 2019, 9, 644–649. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]

| Case Description | ||
|---|---|---|
| Frequency * | Percentage | |
| General features | ||
| Publication Date | ||
| 2009–2011 | 5 | 2.8 |
| 2012–2014 | 24 | 13.6 |
| 2015–2017 | 53 | 30 |
| 2018–2020 | 65 | 36.7 |
| 2021–2022 | 30 | 16.9 |
| Total | 177 | 100 |
| Country | ||
| Brazil | 2 | 1.1 |
| Bulgaria | 4 | 2.3 |
| China | 153 | 86.4 |
| Greece | 1 | 0.6 |
| Egypt | 1 | 0.6 |
| Italy | 4 | 2.3 |
| Japan | 1 | 0.6 |
| Poland | 5 | 2.8 |
| Turkey | 6 | 3.3 |
| Total | 177 | 100 |
| Study type | ||
| Prospective | 5 | 2.8 |
| Retrospective | 172 | 97.2 |
| Total | 177 | 100 |
| Site of miRNA extraction | ||
| Cancer tissue | 160 | 90.4 |
| Peripheral blood | 11 | 6.2 |
| Both | 6 | 3.4 |
| Total | 177 | 100 |
| Blood Compartment | ||
| Serum | 7 | 43.8 |
| Plasma | 6 | 37.5 |
| Extracellular vesicles (EVs) | 2 | 12.5 |
| Missing (Peripheral blood) | 1 | 6.2 |
| Total | 16 | 100 |
| Tumor sampling site | ||
| Superficial and deep | 8 | 4.5 |
| Deep | 2 | 1.1 |
| Not defined | 167 | 94.4 |
| Total | 177 | 100 |
| Tumor features | ||
| T Stage | ||
| T1–T2(early) | 3 | 1.7 |
| T3–T4 (advanced) | 12 | 6.8 |
| T1–T4 (any stage) | 99 | 55.9 |
| Not defined | 63 | 35.6 |
| Total | 177 | 100 |
| Localization (laryngeal subsites) | ||
| All sites | 29 | 16.4 |
| Supraglottic | 5 | 2.8 |
| Supraglottic and glottic | 20 | 11.3 |
| Glottic | 1 | 0.6 |
| Not defined | 122 | 68.9 |
| Total | 177 | 100 |
| Control Group | ||
| Normal laryngeal mucosa (same patient) | 143 | 80.8 |
| Normal laryngeal mucosa (same patient) and healthy blood (another person) | 4 | 2.3 |
| Normal laryngeal mucosa (another patient) | 13 | 7.3 |
| Other (including healthy blood from another patient) | 15 | 8.5 |
| Not defined | 2 | 1.1 |
| Total | 177 | 100 |
| miRNA Description | ||
|---|---|---|
| Frequency * | Percentage | |
| Type of microRNA | ||
| Tissue miRNA | 182 | 78.4 |
| Circulating miRNA | 50 | 21.6 |
| Total | 232 | 100 |
| Tissue miRNA | ||
| Described by more than 10 papers | 2 | 1.1 |
| Described by 5–9 papers | 9 | 4.9 |
| Described by 2–4 papers | 54 | 29.7 |
| Described by only 1 paper | 117 | 64.3 |
| Total | 182 | 100 |
| Circulating miRNA | ||
| Described by 2–3 papers | 7 | 14 |
| Described by only 1 paper | 43 | 86 |
| Total | 50 | 100 |
| Circulating miRNA | Dysplasia | LSCC | Grading | T stage | N Stage | Number of Paper(s) * | Cohort Numerosity (Range) # | Ref(s) |
|---|---|---|---|---|---|---|---|---|
| Downregulated | ||||||||
| miR-125b-5p | 0 | 2 ↓ | 0 | 0 | 0 | 2 | 124 (60–64) | [101,133] |
| miR-126-3p | 0 | 2 ↓ | 1↓ | 0 | 0 | 2 | 102 (38–64) | [133,141] |
| miR-133a-3p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-145-5p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-150-5p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-19a-3p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-192-5p | 0 | 1↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-203-3p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-218-5p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-25-3p | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-451a | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-601 | 0 | 1 ↓ | 0 | 0 | 0 | 1 | 64 | [133] |
| Upregulated | ||||||||
| let-7a-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-106b-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-10a-5p | 0 | 1 ↑ | 1 ↑ | 1 ↑ | 0 | 1 | 236 | [57] |
| miR-122-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-1246 | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 61 | [61] |
| miR-130a-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-141-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-149a-5p | 0 | 1 ↑ | 1 ↑ | 1 ↑ | 1 ↑ | 1 | 66 | [79] |
| miR-155-5p | 0 | 1 ↑ | 0 | 1 ↑ | 1 ↑ | 1 | 840 | [169] |
| miR-182-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-191-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-195-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-196a-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 22 | [39] |
| miR-19b-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-20a-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-21-5p | 1 ↑ | 3 ↑ | 0 | 2 ↑ | 1 ↑ | 3 | 404 (52–236) | [57,143,174] |
| miR-221-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 60 | [154] |
| miR-26b-5p | 0 | 1 ↑ | 0 | 1 ↑ | 0 | 1 | 59 | [116] |
| miR-27a-3p | 0 | 2 ↑ | 0 | 0 | 0 | 2 | 128 (64–64) | [133,191] |
| miR-27b-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-30b-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-31-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 22 | [39] |
| miR-31-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-320a-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-328-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-33a-5p | 0 | 2 ↑ | 0 | 0 | 0 | 2 | 76 (10–66) | [79,140] |
| miR-375-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-378a-5p | 0 | 1 ↑ | 1 ↑ | 1 ↑ | 0 | 1 | 384 | [189] |
| miR-424-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 22 | [39] |
| miR-484 | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-485-3p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 66 | [79] |
| miR-632 | 0 | 1 ↑ | 1↑ | 1↑ | 1↑ | 1 | 162 | [63] |
| miR-93-5p | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 64 | [133] |
| miR-941 | 0 | 1 ↑ | 0 | 0 | 0 | 1 | 59 | [33] |
| Other | ||||||||
| miR-206 | 0 | 1 ↑,1 ↓ | 0 | 0 | 0 | 2 | 64↑ 68↓ | [133,193] |
| miR-223-3p | 0 | 1 ↑,1 ↓ | 0 | 0 | 0 | 2 | 64↑ 66↓ | [79,133] |
| Tissue miRNA | Recurrence | Response to RT | Response to CHT | DSS | Number of Paper(s) * | Cohort Numerosity (Range) # | Ref(s) |
|---|---|---|---|---|---|---|---|
| Tumor suppressor miRNA | |||||||
| miR-101-3p | 0 | 0 | 0 | 1 | 1 | 80 | [156] |
| miR-107 | 0 | 0 | 1 | 0 | 1 | 30 | [22] |
| miR-1205 | 1 | 0 | 0 | 1 | 1 | 44 | [70] |
| miR-140-5p | 0 | 0 | 0 | 1 | 1 | 56 | [83] |
| miR-143-3p | 0 | 0 | 0 | 2 | 2 | 112 (52–60) | [90,93] |
| miR-145-5p | 0 | 0 | 0 | 2 | 2 | 320 (132–188) | [92,99] |
| miR-147a | 0 | 0 | 0 | 1 | 1 | 45 | [24] |
| miR-149-5p | 0 | 0 | 0 | 1 | 1 | 143 | [179] |
| miR-154-5p | 0 | 0 | 0 | 1 | 1 | 104 | [100] |
| miR-195-5p | 1 | 0 | 0 | 3 | 3 | 402 (98–182) | [114,115,197] |
| miR-204-5p | 0 | 0 | 0 | 1 | 1 | 20 | [49] |
| miR-22-3p | 1 | 0 | 0 | 1 | 1 | 49 | [76] |
| miR-29c-3p | 0 | 0 | 0 | 1 | 1 | 66 | [74] |
| miR-300 | 0 | 0 | 0 | 1 | 1 | 133 | [190] |
| miR-375-3p | 0 | 0 | 0 | 1 | 1 | 46 | [159] |
| miR-497-5p | 0 | 0 | 1 | 0 | 1 | 38 | [30] |
| miR-518a-3p | 0 | 0 | 1 | 0 | 1 | 60 | [47] |
| miR-519a-3p | 0 | 0 | 0 | 1 | 1 | 96 | [135] |
| miR-655-3p | 0 | 0 | 1 | 0 | 1 | 105 | [23] |
| miR-766-5p | 0 | 0 | 0 | 1 | 1 | 60 | [40] |
| miR-873-5p | 0 | 0 | 1 | 0 | 1 | 28 | [35] |
| OncomiRs | |||||||
| miR-1246 | 0 | 0 | 0 | 1 | 1 | 61 | [61] |
| miR-144-3p | 1 | 0 | 0 | 0 | 1 | 60 | [42] |
| miR-146a-5p | 0 | 0 | 0 | 1 | 1 | 33 | [50] |
| miR-17-5p | 0 | 0 | 0 | 1 | 1 | 39 | [66] |
| miR-196b-3p | 0 | 0 | 0 | 1 | 1 | 79 | [91] |
| miR-196b-5p | 0 | 0 | 0 | 1 | 1 | 113 | [103] |
| miR-19a-3p | 0 | 0 | 0 | 1 | 1 | 83 | [142] |
| miR-20b-5p | 0 | 0 | 0 | 1 | 1 | 105 | [31] |
| miR-210-3p | 0 | 0 | 0 | 1 | 1 | 60 | [42] |
| miR-21-5p | 0 | 0 | 0 | 2 | 2 | 92 (46–46) | [147,159] |
| miR-23a-3p | 0 | 0 | 0 | 1 | 1 | 52 | [155] |
| miR-26a-5p | 0 | 0 | 0 | 1 | 1 | 56 | [53] |
| miR-27a-3p | 0 | 0 | 0 | 1 | 1 | 62 | [191] |
| miR-296-5p | 1 | 1 | 0 | 0 | 1 | 34 | [157] |
| miR-301a-3p | 0 | 0 | 0 | 1 | 1 | 120 | [163] |
| miR-34b-5p | 0 | 0 | 0 | 1 | 1 | 33 | [50] |
| miR-34c-5p | 2 | 0 | 0 | 2 | 2 | 133 (43–90) | [110,151] |
| miR-93-5p | 0 | 0 | 0 | 1 | 1 | 60 | [42] |
| miR-9-5p | 0 | 0 | 0 | 1 | 1 | 103 | [150] |
| Circulating miRNA | Recurrence | DSS | Number of Paper(s) * | Cohort Numerosity # | Ref(s) |
|---|---|---|---|---|---|
| OncomiR | |||||
| miR-1246 | 0 | 1 | 1 | 61 | [61] |
| miR-21-5p | 0 | 1 | 1 | 236 | [57] |
| miR-26b-5p | 1 | 1 | 1 | 59 | [116] |
| miR-632 | 1 | 1 | 1 | 162 | [63] |
| Tumor suppressor miRNA | |||||
| miR-10a-5p | 0 | 1 | 1 | 236 | [57] |
| miR-126-3p | 0 | 1 | 1 | 38 | [141] |
| miR-378a-3p | 1 | 0 | 1 | 384 | [189] |
| miRNA Functional Analysis Papers Description | |||
|---|---|---|---|
| OncomiRs | Tumor Suppressor miRNAs | Total | |
| Papers | 12 * | 44 * | 56 * |
| miRNAs | 12 (26.1%) | 34 (73.9%) | 46 (100%) |
| In vitro models | |||
| 1 LSCC cell line | 7 (12.5%) | 24 (42.9%) | 31 (55.4%) |
| 2 LSCC cell lines | 4 (7.1%) | 15 (26.8%) | 19 (33.9.%) |
| 3 LSCC cell lines | 1 (1.8%) | 4 (7.1%) | 5 (8.9%) |
| 4 LSCC cell lines | - | 1 (1.8%) | 1 (1.8%) |
| Total | 12 (21.4%) | 44 (78.6%) | 56 (100%) |
| LSCC cell lines | |||
| AMC-HN-8 | 3 (3.4%) | 20 (22.7%) | 23 (26.1%) |
| TU-177 | 4 (4.6%) | 19 (21.6%) | 23 (26.2%) |
| TU686 | 3 (3.4%) | 11 (12.5%) | 14 (15.9%) |
| SNU889 | 3 (3.4%) | 5 (5.7%) | 8 (9.1%) |
| Others | 5 (5.7%) | 15 (17%) | 20 (22.7%) |
| Total | 18 (20.5%) | 70 (79.5%) | 88 (100%) |
| Validated Target | |||
| Yes | 8 (17.4%) | 29 (63%) | 37 (80.4%) |
| No | 4 (8.7%) | 5 (10.9%) | 9 (19.6%) |
| Total | 12 (26.1%) | 34 (73.9%) | 46 (100%) |
| Cellular processes | |||
| Cell proliferation and cycle | 7 (6.3%) | 38 (34.6%) | 41 (40.9%) |
| Apoptosis | 1 (0.9%) | 18 (16.4%) | 19 (17.3%) |
| Invasion and migration | 9 (8.2%) | 30 (27.3%) | 39 (35.5%) |
| Metabolism | - | 1 (0.9%) | 1 (0.9%) |
| Drug sensitive | - | 3 (2.7%) | 3 (2.7%) |
| NA | 3 (2.7%) | 3 (2.7%) | |
| Total | 17 (15.4%) | 93 (84.6%) | 110 (100%) |
| miRNA | LSCC cell line(s) | Target(s) | Cellular Processes | Ref(s) |
|---|---|---|---|---|
| OncomiRs | ||||
| miR-148a-3p | TU686 | DNMT1 | Proliferation, invasion and migration | [182] |
| miR-155-5p | AMC-HN-8, TU-177 | - | Proliferation, invasion and migration | [84] |
| miR-196a-5p | JHU-011 | - | Proliferation, tumor growth and metastasis | [131] |
| miR-205-5p | SNU899 | - | Invasion and migration | [188] |
| miR-27a-3p | AMC-HN-8, TU686 | SMAD4 | Proliferation, invasion and migration, tumor growth | [191] |
| miR-301a-3p | TU-177 | SMAD4 | Proliferation, apoptosis, invasion and migration | [163] |
| miR-302b-3p | SNU899, SNU1066, SNU1076 | TGFBR2 | Invasion and migration | [52] |
| miR-340-3p | TU-177 | ELK1 | - | [38] |
| miR-503-5p | AMC-HN-8 | PDCD4 | Proliferation, invasion and migration | [112] |
| miR-744-3p | SNU899, SNU1076, | PDCD4, PTEN | Invasion and migration | [177] |
| miR-941 | TU686, FD-LSC-1 | - | Proliferation, invasion and migration | [33] |
| miR-98-5p | TU-177 | HMGA2 | - | [77] |
| Tumor suppressor miRNAs | ||||
| let-7c-5p | TU-177, FD-LSC-1 | - | Proliferation, apoptosis, invasion and migration | [34] |
| miR-107 | TU686 | CACNA2D1 | Proliferation, invasion and migration | [72] |
| miR-125b-5p | AMC-HN-8, TU-177 | STAT3, HK2 | Proliferation, invasion and migration, metabolism | [101,117] |
| miR-136a-5p | TU686 | RAP2C | - | [64] |
| miR-139-3p | TU-177, HNO210, | KDM5B | Proliferation, apoptosis, migration and invasion | [36] |
| miR-140-5p | AMC-HN-8, TU-177, SNU46, UM-SCC-10B | FGF9, NFAT5 | Proliferation, apoptosis, migration and invasion | [68,192] |
| miR-141-3p | AMC-HN-8 | HOXC6 | Proliferation, migration and invasion, tumor growth and metastasis | [73] |
| miR-143-3p | TU-177, TU686, SNU889 | k-Ras, MAGE-A9 | Proliferation, apoptosis, invasion and migration, tumor growth | [90,93] |
| miR-145-5p | TU-177 | FSCN1 | Proliferation and cycle, apoptosis, invasion and migration | [92] |
| miR-195-5p | AMC-HN-8, TU-177 | cyclin D1, ROCK1, DCUN1D1 | Proliferation and cycle, apoptosis, migration and invasion | [75,107,111,114] |
| miR-204-5p | TU-177, AMC-HN-3, HN-10 | ZEB1, FOXC1, SEMA4B | Proliferation, apoptosis, migration and invasion, tumor growth | [49,65,113] |
| miR-206 | SNU899, TR-LCC-1 | SOX9 | Proliferation, apoptosis, migration and invasion | [193] |
| miR-218-5p | AMC-HN-8 | CCAT1 | Proliferation, migration and invasion | [118] |
| miR-22-3p | AMC-HN-8 | NLRP3 | Migration and invasion | [76] |
| miR-24-3p | AMC-HN-8 | XIAP | Proliferation, apoptosis, response to treatment (radiosensitivity) | [152] |
| miR-330-3p | AMC–HN–8, TU686, SNU-899, FD-LSC-1 | Tra2β, SLC7A11 | Proliferation, apoptosis, invasion and migration | [56,58] |
| miR-340–5p | AMC-HN-8, TU177, TU686 | NEAT1 | Proliferation, invasion and migration, tumor growth | [54] |
| miR-363-3p | TU-177 | Mcl-1 | Proliferation, apoptosis, invasion and migration | [98] |
| miR-365a-3p | UM-SCC-17A | - | Proliferation | [44] |
| miR-375-3p | SNU899, SNU46 | IGF1R | Proliferation, apoptosis, invasion and migration | [149,188] |
| miR-381-3p | AMC-HN-3 | NASP | Proliferation, invasion and migration | [71] |
| miR-384 | TU686 | WISP1 | Proliferation, apoptosis | [89] |
| miR-449a | HNO210 | - | Proliferation | [28] |
| miR-4497 | TU686, UM-SCC-17A | GBX2 | Proliferation, apoptosis, migration and invasion | [102] |
| miR-4735-3p | AMC-HN-8, TU-177, UM-SCC-17A | TNFAIP3 | Proliferation, invasion and migration | [51] |
| miR-486-3p | TU686, AMC-HN-8 | LASP1 | Proliferation and cycle, apoptosis, migration and invasion | [37] |
| miR-493-3p | AMC-HN-8 | - | Proliferation, apoptosis | [69] |
| miR-506-3p | TU-177 | YAP1 | Proliferation and cycle, apoptosis, migration and invasion | [87] |
| miR-518a-3p | TU-177, TU686 | SPATS2L | Response to treatment (cisplatin sensitivity) | [47] |
| miR-613 | TU686 | PDK1 | Proliferation, invasion and migration | [109] |
| miR-625-5p | AMC-HN-8, TU-177 | SOX4 | Proliferation, invasion and migration | [88] |
| miR-654-3p | TU-177, FD-LSC-1 | USP7 | - | [55] |
| miR-766-5p | AMC-HN-8, TU686 | HMGA2 | Invasion and migration | [40] |
| miR-873-5p | AMC-HN-8, TU-177 | - | Proliferation, apoptosis, migration and invasion, response to treatment (cisplatin sensitivity) | [35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broseghini, E.; Filippini, D.M.; Fabbri, L.; Leonardi, R.; Abeshi, A.; Dal Molin, D.; Fermi, M.; Ferracin, M.; Fernandez, I.J. Diagnostic and Prognostic Value of microRNAs in Patients with Laryngeal Cancer: A Systematic Review. Non-Coding RNA 2023, 9, 9. https://doi.org/10.3390/ncrna9010009
Broseghini E, Filippini DM, Fabbri L, Leonardi R, Abeshi A, Dal Molin D, Fermi M, Ferracin M, Fernandez IJ. Diagnostic and Prognostic Value of microRNAs in Patients with Laryngeal Cancer: A Systematic Review. Non-Coding RNA. 2023; 9(1):9. https://doi.org/10.3390/ncrna9010009
Chicago/Turabian StyleBroseghini, Elisabetta, Daria Maria Filippini, Laura Fabbri, Roberta Leonardi, Andi Abeshi, Davide Dal Molin, Matteo Fermi, Manuela Ferracin, and Ignacio Javier Fernandez. 2023. "Diagnostic and Prognostic Value of microRNAs in Patients with Laryngeal Cancer: A Systematic Review" Non-Coding RNA 9, no. 1: 9. https://doi.org/10.3390/ncrna9010009