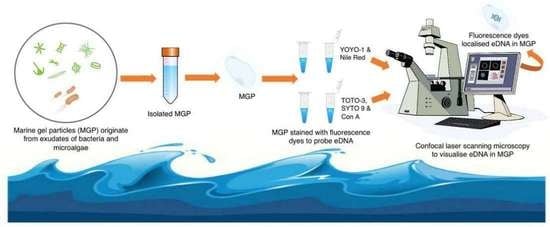
Isolation and Staining Reveal the Presence of Extracellular DNA in Marine Gel Particles
Abstract
:1. Introduction
2. Results
2.1. Microscopy of Purified Natural MGPs in Liquid Suspension
2.2. Visualisation of the Natural MGP Matrix
2.3. Visualisation of eDNA in Natural MGP Matrix
2.4. A Pseudoalteromonas atlantica MGP Model
2.5. Visualisation of the P. atlantica MGP Model Matrix
2.6. Visualisation of eDNA in P. atlantica EPS (MGP Model)
3. Discussion
3.1. Isolation of MGPs in Liquid Suspension
3.2. The Presence of eDNA in Natural MGPs and P. atlantica MGPs
4. Conclusions
5. Materials and Methods
5.1. Seawater Collection
5.2. Isolation of Natural MGPs from Seawater by Filtration
5.3. Isolation of Concentrated Natural MGPs in Suspension
5.4. Bacterial Strains and Growth Conditions
5.5. A Model MGP Using Pseudoalteromonas atlantica
5.6. Compositional Analysis of Natural MGPs and a P. atlantica MGP Model
5.7. MGP Staining with eDNA Probes
5.8. MGP Visualisation by Microscopy
5.9. Bioimaging
5.10. Quantification of Fluorescent Dye Intensity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quigg, A.; Santschi, P.H.; Burd, A.; Chin, W.-C.; Kamalanathan, M.; Xu, C.; Ziervogel, K. From Nano-Gels to Marine Snow: A Synthesis of Gel Formation Processes and Modeling Efforts Involved with Particle Flux in the Ocean. Gels 2021, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Thornton, D. Transparent exopolymer particle production and aggregation by a marine planktonic diatom (Thalassiosira weissflogii) at different growth rates. J. Phycol. 2015, 51, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Gutierrez, T. Microbial Extracellular Polymeric Substances (EPSs) in Ocean Systems. Front. Microbiol. 2017, 8, 922. [Google Scholar] [CrossRef] [PubMed]
- Passow, U. Transparent exopolymer particles (TEP) in aquatic environments. Prog. Oceanogr. 2002, 55, 287–333. [Google Scholar] [CrossRef] [Green Version]
- Chin, W.-C.; Orellana, M.; Verdugo, P. Spontaneous assembly of marine dissolved organic matter into polymer gels. Nature 1998, 391, 568–572. [Google Scholar] [CrossRef]
- Verdugo, P.; Alldredge, A.L.; Azam, F.; Kirchman, D.L.; Passow, U.; Santschi, P.H. The oceanic gel phase: A bridge in the DOM–POM continuum. Mar. Chem. 2004, 92, 67–85. [Google Scholar] [CrossRef]
- Verdugo, P.; Santschi, P. Polymer dynamics of DOC networks and gel formation in seawater. Deep Sea Res. Part II Top. Stud. Oceanogr. 2010, 57, 1486–1493. [Google Scholar] [CrossRef]
- Mari, X.; Passow, U.; Migon, C.; Burd, A.B.; Legendre, L. Transparent exopolymer particles: Effects on carbon cycling in the ocean. Prog. Oceanogr. 2017, 151, 13–37. [Google Scholar] [CrossRef] [Green Version]
- Christina, L.; Passow, U. Factors influencing the sinking of POC and the efficiency of the biological carbon pump. Deep Sea Res. Part II Top. Stud. Oceanogr. 2007, 54, 639–658. [Google Scholar]
- Shanks, A.L. The abundance, vertical flux, and still-water and apparent sinking rates of marine snow in a shallow coastal water column. Cont. Shelf Res. 2002, 22, 2045–2064. [Google Scholar] [CrossRef]
- Kiørboe, T. Formation and fate of marine snow: Small-scale processes with large-scale implications. Sci. Mar. 2001, 65, 57–71. [Google Scholar] [CrossRef]
- Li, S.; Winters, H.; Villacorte, L.O.; Ekowati, Y.; Emwas, A.H.; Kennedy, M.D.; Amy, G.L. Compositional similarities and differences between transparent exopolymer particles (TEPs) from two marine bacteria and two marine algae: Significance to surface biofouling. Mar. Chem. 2015, 174, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Park, C.; Kim, H.; Park, H.; Hong, S. Role of transparent exopolymer particles (TEP) in initial bacterial deposition and biofilm formation on reverse osmosis (RO) membrane. J. Membr. Sci. 2015, 494, 25–31. [Google Scholar] [CrossRef]
- Bar-Zeev, E.; Berman-Frank, I.; Liberman, B.; Rahav, E.; Passow, U.; Berman, T. Transparent exopolymer particles: Potential agents for organic fouling and biofilm formation in desalination and water treatment plants. Desalination Water Treat. 2009, 3, 136–142. [Google Scholar] [CrossRef] [Green Version]
- Bar-Zeev, E.; Berman-Frank, I.; Girshevitz, O.; Berman, T. Revised paradigm of aquatic biofilm formation facilitated by microgel transparent exopolymer particles. Proc. Natl. Acad. Sci. USA 2012, 109, 9119–9124. [Google Scholar] [CrossRef] [Green Version]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Deng, B.; Ghatak, S.; Sarkar, S.; Ghatak, P.; McComb, D.W.; Sen, C.K. STEM Observation of eDNA as a Dominant Component of EPS in Pseudomonas aeruginosa Biofilm. Microsc. Microanal. 2018, 24, 1334–1335. [Google Scholar] [CrossRef] [Green Version]
- Okshevsky, M.; Meyer, R. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef]
- Okshevsky, M.; Regina, V.; Meyer, R. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef]
- Pakkulnan, R.; Anutrakunchai, C.; Kanthawong, S.; Taweechaisupapong, S.; Chareonsudjai, P.; Chareonsudjai, S. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PloS ONE 2019, 14, e0213288. [Google Scholar] [CrossRef]
- Alldredge, A.L.; Passow, U.; Logan, B. The Abundance and Significance of a Class of Large, Transparent Organic Particles in the Ocean. Deep-Sea Res. Part I-Oceanogr. Res. Pap. 1993, 40, 1131–1140. [Google Scholar] [CrossRef]
- Alldredge, A.L.; Silver, M. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 1988, 20, 41–82. [Google Scholar] [CrossRef]
- Busch, K.; Endres, S.; Iversen, M.H.; Michels, J.; Nöthig, E.M.; Engel, A. Bacterial colonization and vertical distribution of marine gel particles (TEP and CSP) in the Arctic Fram Strait. Front. Mar. Sci. 2017, 4, 166. [Google Scholar] [CrossRef] [Green Version]
- Thornton, D.C.O. Coomassie Stainable Particles (CSP): Protein Containing Exopolymer Particles in the Ocean. Front. Mar. Sci. 2018, 5, 206. [Google Scholar] [CrossRef]
- Cisternas-Novoa, C.; Lee, C.; Engel, A. Transparent exopolymer particles (TEP) and Coomassie stainable particles (CSP): Differences between their origin and vertical distributions in the ocean. Mar. Chem. 2015, 175, 56–71. [Google Scholar] [CrossRef]
- Ortega-Retuerta, E.; Duarte, C.; Reche, I. Significance of bacterial activity for the distribution and dynamics of transparent exopolymer particles in the Mediterranean Sea. Microb. Ecol. 2010, 59, 808–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Cruz, B.; Neuer, S. Effects of nutrient limitation on cell growth, TEP production and aggregate formation of marine Synechococcus. Aquat. Microb. Ecol. 2016, 78, 39–49. [Google Scholar] [CrossRef]
- Iuculano, F.; Duarte, C.M.; Marbà, N.; Agustí, S. Seagrass as major source of transparent exopolymer particles in the oligotrophic Mediterranean coast. Biogeosciences 2017, 14, 5069–5075. [Google Scholar] [CrossRef] [Green Version]
- Stoderegger, K.E.; Herndl, G. Production of exopolymer particles by marine bacterioplankton under contrasting turbulence conditions. Mar. Ecol. Prog. Ser. 1999, 189, 9–16. [Google Scholar] [CrossRef]
- Passow, U. Production of transparent exopolymer particles (TEP) by phyto-and bacterioplankton. Mar. Ecol. Prog. Ser. 2002, 236, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hewes, C.D.; Holm-Hansen, O. A method for recovering nanoplankton from filters for identification with the microscope: The filter-transfer-freeze (FTF) technique 1. Limnol. Oceanogr. 1983, 28, 389–394. [Google Scholar] [CrossRef]
- Engel, A. Determination of Marine Gel Particles. In Practical Guidelines for the Analysis of Seawater; CRC Press: Boca Raton, FL, USA, 2009; pp. 125–142. [Google Scholar]
- Villacorte, L.O.; Ekowati, Y.; Calix-Ponce, H.N.; Schippers, J.C.; Amy, G.L.; Kennedy, M.D. Improved method for measuring transparent exopolymer particles (TEP) and their precursors in fresh and saline water. Water Res. 2015, 70, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Thuy, N.T.; Huang, C.-P.; Lin, J.-L. Visualization and quantification of transparent exopolymer particles (TEP) in freshwater using an auto-imaging approach. Environ. Sci. Pollut. Res. 2017, 24, 17358–17372. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sharma, P.K.; Busscher, H.J.; Van Der Mei, H.C.; Krom, B.P. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef] [Green Version]
- Okshevsky, M.; Meyer, R. Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms. J. Microbiol. Methods 2014, 105, 102–104. [Google Scholar] [CrossRef]
- Yuan, B.; Wang, X.; Tang, C.; Li, X.; Yu, G. In situ observation of the growth of biofouling layer in osmotic membrane bioreactors by multiple fluorescence labeling and confocal laser scanning microscopy. Water Res. 2015, 75, 188–200. [Google Scholar] [CrossRef]
- Reichhardt, C.; Parsek, M. Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 2019, 10, 677. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Neu, T.R.; Zhang, Y.; Bellenberg, S.; Kuhlicke, U.; Li, Q.; Sand, W.; Vera, M. Visualization and analysis of EPS glycoconjugates of the thermoacidophilic archaeon Sulfolobus metallicus. Appl. Microbiol. Biotechnol. 2015, 99, 7343–7356. [Google Scholar] [CrossRef]
- Schlafer, S.; Meyer, R. Confocal microscopy imaging of the biofilm matrix. J. Microbiol. Methods 2017, 138, 50–59. [Google Scholar] [CrossRef]
- Cowen, J.; Holloway, C. Structural and chemical analysis of marine aggragates: In situ macrophotography and laser confocal and electron microscopy. Mar. Biol. 1996, 126, 163–174. [Google Scholar] [CrossRef]
- Holloway, C.F.; Cowen, J. Development of a scanning confocal laser microscopic technique to examine the structure and composition of marine snow. Limnol. Oceanogr. 1997, 42, 1340–1352. [Google Scholar] [CrossRef] [Green Version]
- Flintrop, C.M.; Rogge, A.; Miksch, S.; Thiele, S.; Waite, A.M.; Iversen, M.H. Embedding and slicing of intact in situ collected marine snow. Limnol. Oceanogr. Methods 2018, 16, 339–355. [Google Scholar] [CrossRef] [Green Version]
- Samo, T.J.; Malfatti, F.; Azam, F. A new class of transparent organic particles in seawater visualized by a novel fluorescence approach. Aquat. Microb. Ecol. 2008, 53, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Li, L.; Sharma, S.; Wang, J.; McHardy, I.; Lux, R.; Yang, Z.; He, X.; Gimzewski, J.K.; Li, Y.; et al. DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS ONE 2012, 7, e51905. [Google Scholar] [CrossRef]
- Qin, Z.; Ou, Y.; Yang, L.; Zhu, Y.; Tolker-Nielsen, T.; Molin, S.; Qu, D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 2007, 153, 2083–2092. [Google Scholar] [CrossRef] [Green Version]
- Gallo, P.M.; Rapsinski, G.J.; Wilson, R.P.; Oppong, G.O.; Sriram, U.; Goulian, M.; Buttaro, B.; Caricchio, R.; Gallucci, S.; Tükel, Ç. Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 2015, 42, 1171–1184. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Winters, H.; Jeong, S.; Emwas, A.H.; Vigneswaran, S.; Amy, G.L. Marine bacterial transparent exopolymer particles (TEP) and TEP precursors: Characterization and RO fouling potential. Desalination 2016, 379, 68–74. [Google Scholar] [CrossRef]
- Yamada, Y.; Fukuda, H.; Tada, Y.; Kogure, K.; Nagata, T. Bacterial enhancement of gel particle coagulation in seawater. Aquat. Microb. Ecol. 2016, 77, 11–22. [Google Scholar] [CrossRef] [Green Version]
- McDonnell, A.M.; Buesseler, K. Variability in the average sinking velocity of marine particles. Limnol. Oceanogr. 2010, 55, 2085–2096. [Google Scholar] [CrossRef]
- Ploeger, L.; Dullens, H.F.J.; Huisman, A.; Van Diest, P.J. Fluorescent stains for quantification of DNA by confocal laser scanning microscopy in 3-D. Biotech. Histochem. 2008, 83, 63–69. [Google Scholar] [CrossRef]
- Arruda Fatibello, S.H.S.; Vieira, A.H.; Fatibello-Filho, O. A rapid spectrophotometric method for the determination of transparent exopolymer particles (TEP) in freshwater. Talanta 2004, 62, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Discart, V. Transparent Exopolymer Particles: Detection and Role in Membrane Based Systems. Ph.D. Thesis, KU Leuven, Leuven, Belgium, 2015. [Google Scholar]
- Cisternas-Novoa, C.; Lee, C.; Engel, A. A semi-quantitative spectrophotometric, dye-binding assay for determination of Coomassie Blue stainable particles. Limnol. Oceanogr. Methods 2014, 12, 604–616. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Chin, W.C.; Lin, P.; Chen, H.; Chiu, M.H.; Waggoner, D.C.; Xing, W.; Sun, L.; Schwehr, K.A.; Hatcher, P.G.; et al. Comparison of microgels, extracellular polymeric substances (EPS) and transparent exopolymeric particles (TEP) determined in seawater with and without oil. Mar. Chem. 2019, 215, 103667. [Google Scholar] [CrossRef]
- Serrage, H.; Jepson, M.A.; Rostami, N.; Jakubovics, N.S.; Nobbs, A.H. Understanding the Matrix: The Role of Extracellular DNA in Oral Biofilms. Front. Oral. Health 2021, 2, 640129. [Google Scholar] [CrossRef]
- Olsen, A.; Ehrhardt, C.; Yadavalli, V. Nanoscale visualization of extracellular DNA on cell surfaces. Anal. Sci. Adv. 2020, 1, 194–202. [Google Scholar] [CrossRef]
- Böckelmann, U.; Manz, W.; Neu, T.R.; Szewzyk, U. Investigation of lotic microbial aggregates by a combined technique of fluorescent in situ hybridization and lectin-binding-analysis. J. Microbiol. Methods 2002, 49, 75–87. [Google Scholar] [CrossRef]
- Thill, A.; Veerapaneni, S.; Simon, B.; Wiesner, M.; Bottero, J.Y.; Snidaro, D. Determination of structure of aggregates by confocal scanning laser microscopy. J. Colloid Interface Sci. 1998, 204, 357–362. [Google Scholar] [CrossRef]
- Waite, A.M.; Safi, K.A.; Hall, J.A.; Nodder, S.D. Mass sedimentation of picoplankton embedded in organic aggregates. Limnol. Oceanogr. 2000, 45, 87–97. [Google Scholar] [CrossRef]
- Neu, T.R. In situ cell and glycoconjugate distribution in river snow studied by confocal laser scanning microscopy. Aquat. Microb. Ecol. 2000, 21, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Seymour, M.; Wilgar, G.; Carvalho, G.R.; De Bruyn, M.; Edwards, F. Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun. Biol. 2018, 1, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Best, S.; Sorensen, P. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Ishikawa, K.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin compaction protects genomic DNA from radiation damage. PLoS ONE 2013, 8, e75622. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Lo, L.; Maier, R. A histone-like protein of Helicobacter pylori protects DNA from stress damage and aids host colonization. DNA Repair 2012, 11, 733–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almarza, O.; Núñez, D.; Toledo, H. The DNA-Binding Protein HU has a Regulatory Role in the Acid Stress Response Mechanism in H elicobacter pylori. Helicobacter 2015, 20, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Salter, I. Seasonal variability in the persistence of dissolved environmental DNA (eDNA) in a marine system: The role of microbial nutrient limitation. PLoS ONE 2018, 13, e0192409. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z.; Ouellette, M.; Makkay, A.M.; Papke, R.T.; Zhaxybayeva, O. Nutrient supplementation experiments with saltern microbial communities implicate utilization of DNA as a source of phosphorus. ISME J. 2021, 15, 2853–2864. [Google Scholar] [CrossRef]
- Costa, D.; Morán, M.C.; Miguel, M.G.; Lindman, B. Cross-Linked DNA Gels and Gel Particles. In DNA Interactions with Polymers and Surfactants; Wiley: Hoboken, NJ, USA, 2008; pp. 353–365. [Google Scholar]
- Lindman, B.; Dias, R.S.; Miguel, M.G.; Morán, M.C.; Costa, D. Manipulation of DNA by Surfactants. In Highlights in Colloid Science; Wiley: Hoboken, NJ, USA, 2008; pp. 179–202. [Google Scholar]
- Tolentino, M.Q.; Hartmann, A.K.; Loe, D.T.; Rouge, J.L. Controlled release of small molecules and proteins from DNA-surfactant stabilized metal organic frameworks. J. Mater. Chem. B 2020, 8, 5627–5635. [Google Scholar] [CrossRef]
- Linney, M.D.; Schvarcz, C.R.; Steward, G.F.; DeLong, E.F.; Karl, D.M. A method for characterizing dissolved DNA and its application to the North Pacific Subtropical Gyre. Limnol. Oceanogr. Methods 2021, 19, 210–221. [Google Scholar] [CrossRef]
- Mukherjee, A.; Sokunbi, A.; Grove, A. DNA protection by histone-like protein HU from the hyperthermophilic eubacterium Thermotoga maritima. Nucleic Acids Res. 2008, 36, 3956–3968. [Google Scholar] [CrossRef] [Green Version]
- Van Holde, K.; Allen, J.R.; Tatchell, K.; Weischet, W.O.; Lohr, D. DNA-histone interactions in nucleosomes. Biophys. J. 1980, 32, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Pal, P.; Modi, M.; Ravichandran, S.; Yennamalli, R.M.; Priyadarshini, R. DNA-binding properties of YbaB, a putative nucleoid-associated protein from Caulobacter crescentus. Front. Microbiol. 2021, 12, 733344. [Google Scholar] [CrossRef]
- Al-Wahaibi, A.S.; Lapinska, E.; Rajarajan, N.; Dobretsov, S.; Upstill-Goddard, R.; Burgess, J.G. Secretion of DNases by marine bacteria: A culture based and bioinformatics approach. Front. Microbiol. 2019, 10, 969. [Google Scholar] [CrossRef] [Green Version]
- Durkin, C.A.; Cetinić, I.; Estapa, M.; Ljubešić, Z.; Mucko, M.; Neeley, A.; Omand, M. Tracing the path of carbon export in the ocean though DNA sequencing of individual sinking particles. ISME J. 2022, 16, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Cruz, B.N.; Brozak, S.; Neuer, S. Microscopy and DNA-based characterization of sinking particles at the Bermuda Atlantic Time-series Study station point to zooplankton mediation of particle flux. Limnol. Oceanogr. 2021, 66, 3697–3713. [Google Scholar] [CrossRef]
- Dyomin, V.; Polovtsev, I.; Davydova, A. Fast Recognition of Marine Particles in Underwater Digital Holography. In Proceeedings of the 23rd International Symposium on Atmospheric and Ocean Optics: Atmospheric Physics, Szczyrk-Gliwice, Poland, 28 February–3 March 2017; International Society for Optics and Photonics: Bellingham, WA, USA, 2017. [Google Scholar]
- Lunven, M.; Landeira, J.M.; Lehaître, M.; Siano, R.; Podeur, C.; Daniélou, M.M.; Le Gall, E.; Gentien, P.; Sourisseau, M. In situ video and fluorescence analysis (VFA) of marine particles: Applications to phytoplankton ecological studies. Limnol. Oceanogr. Methods 2012, 10, 807–823. [Google Scholar] [CrossRef]
- Palomba, E.; Chiaiese, P.; Termolino, P.; Paparo, R.; Filippone, E.; Mazzoleni, S.; Chiusano, M.L. Effects of Extracellular Self- and Nonself-DNA on the Freshwater Microalga Chlamydomonas reinhardtii and on the Marine Microalga Nannochloropsis gaditana. Plants 2022, 11, 1436. [Google Scholar] [CrossRef]
- Al-Wahaibi, A.S.M. Characterisation of Marine Gel Particles and Associated Bacterial Communities. Ph.D. Thesis, Newcastle University, Newcastle, UK, 2020. [Google Scholar]
- Roca, C. Exopolysaccharide production by a marine Pseudoalteromonas sp. strain isolated from Madeira Archipelago ocean sediments. New Biotechnol. 2016, 33, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Passow, U.; Alldredge, A. A dye-binding assay for the spectrophotometric measurement of transparent exopolymer particles (TEP). Limnol. Oceanogr. 1995, 40, 1326–1335. [Google Scholar] [CrossRef]
- Rostami, N.; Shields, R.C.; Yassin, S.A.; Hawkins, A.R.; Bowen, L.; Luo, T.L.; Rickard, A.H.; Holliday, R.; Preshaw, P.M.; Jakubovics, N.S. A Critical Role for Extracellular DNA in Dental Plaque Formation. J. Dent. Res. 2016, 96, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Johnson, Z.I.; Bidigare, R.R.; Blinebry, S.K.; Brown, S.L.; Cullen, J.J.; Loftus, S.E.; Redalje, D.G.; Swink, C.; Van Mooy, B.A. Screening for lipids from marine microalgae using Nile red. Conseq. Microb. Interact. Hydrocarb. Oils Lipids Prod. Fuels Chem. 2017, 1–22. [Google Scholar]
- Dsouza, R.N.; Pischel, U.; Nau, W. Fluorescent dyes and their supramolecular host/guest complexes with macrocycles in aqueous solution. Chem. Rev. 2011, 111, 7941–7980. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Pishko, M.V.; Gefrides, C.C.; McShane, M.J.; Cote, G.L. A fluorescence-based glucose biosensor using concanavalin A and dextran encapsulated in a poly (ethylene glycol) hydrogel. Anal. Chem. 1999, 71, 3126–3132. [Google Scholar] [CrossRef] [PubMed]
- Grabenhorst, L.; Trofymchuk, K.; Steiner, F.; Glembockyte, V.; Tinnefeld, P. Fluorophore photostability and saturation in the hotspot of DNA origami nanoantennas. Methods Appl. Fluoresc. 2020, 8, 024003. [Google Scholar] [CrossRef] [PubMed]













| Fluorophore | Target | Excitation (nm) | Acquisition (nm) |
|---|---|---|---|
| YOYO-1 | eDNA | 491 | 501–532 |
| Nile Red | Cell membrane | 553 | 567–781 |
| Concavalian A (TMR) | Glycoproteins | 552 | 560–643 |
| TOTO-3 | eDNA | 642 | 652–784 |
| SYTO-9 | DNA | 483 | 490–591 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Wahaibi, A.S.M.; Upstill-Goddard, R.C.; Burgess, J.G. Isolation and Staining Reveal the Presence of Extracellular DNA in Marine Gel Particles. Gels 2023, 9, 251. https://doi.org/10.3390/gels9030251
Al-Wahaibi ASM, Upstill-Goddard RC, Burgess JG. Isolation and Staining Reveal the Presence of Extracellular DNA in Marine Gel Particles. Gels. 2023; 9(3):251. https://doi.org/10.3390/gels9030251
Chicago/Turabian StyleAl-Wahaibi, Aisha S. M., Robert C. Upstill-Goddard, and J. Grant Burgess. 2023. "Isolation and Staining Reveal the Presence of Extracellular DNA in Marine Gel Particles" Gels 9, no. 3: 251. https://doi.org/10.3390/gels9030251