Metabolic Engineering of Saccharomyces cerevisiae for Efficient Retinol Synthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Medium and Reagents
2.2. Construction of Plasmids and Strains
2.3. Insertion of Degradation Label
2.4. Structural Simulation and Molecular Docking
2.5. Culture and Fermentation Conditions
2.6. Extraction and Detection Methods
2.7. Statistical Analysis
3. Results and Discussions
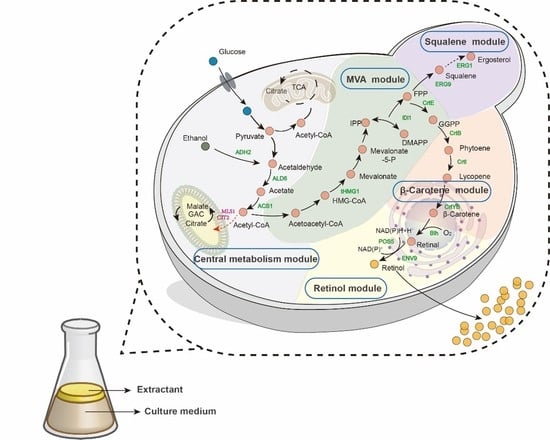
3.1. Construction of the De Novo Retinol Synthesis Pathway in S. cerevisiae
3.2. Module Engineering of the Retinol Metabolism Network
3.3. Knockout of Retinal Transporter
3.4. Screening and Directed Evolution of Retinol Dehydrogenase
3.5. Expansion of the Storage Space for Lipid Droplets
3.6. Improvement of Oxidation Resistance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, E.C.; Choi, J.S.; Joo, C.K. A comparison of vitamin a and cyclosporine a 0.05% eye drops for treatment of dry eye syndrome. Am. J. Ophthalmol. 2009, 147, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Coerdt, K.M.; Goggins, C.A.; Khachemoune, A. Vitamins A, B, C, and D: A short review for the dermatologist. Altern. Ther. Health Med. 2021, 27, 41–49. [Google Scholar] [PubMed]
- Monga, M. Vitamin A and its congeners. Semin. Perinatol. 1997, 27, 135–142. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Laudert, D.; Létinois, U.; Mcclymont, T.; Medlock, J.; Netscher, T. One hundred years of vitamins—A success story of the natural sciences. Angew. Chem. Int. Edit. 2012, 51, 12960–12990. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.L.; Smith, L.K.; Baxendale, I.R. Development of the industrial synthesis of vitamin A. Tetrahedron 2016, 72, 1645–1652. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, M.J.; Canfield, L.M. Enzymatic hydrolysis, extraction, and quantitation of retinol and major carotenoids in mature human milk. J. Nutr. Biochem. 1998, 9, 178–183. [Google Scholar] [CrossRef]
- Manchand, P.S.; Rosenberger, M.; Saucy, G.; Wehrli, P.A.; Wong, H.; Chambers, L.; Ferro, M.P.; Jackson, W. Synthesis of vitamin A via sulfones: A C15 sulfone route. Helv. Chim. Acta 1976, 59, 96–387. [Google Scholar] [CrossRef]
- Anderson, L.A.; Islam, M.A.; Prather, K.L.J. Synthetic biology strategies for improving microbial synthesis of “green” biopolymers. Biol. Chem. 2018, 293, 5053–5061. [Google Scholar] [CrossRef]
- Sun, L.; Kwak, S.; Jin, Y.S. Vitamin A production by engineered Saccharomyces cerevisiae from Xylose via two-phase in situ extraction. ACS Synth. Biol. 2019, 8, 2131–2140. [Google Scholar] [CrossRef]
- Lee, Y.G.; Kim, C.; Sun, L.; Lee, T.H.; Jin, Y.S. Selective production of retinol by engineered Saccharomyces cerevisiae through the expression of retinol-dehydrogenase. Biotechnol. Bioeng. 2022, 119, 399–410. [Google Scholar] [CrossRef]
- Park, H.; Lee, D.; Kim, J.E.; Park, S.; Park, J.H.; Ha, C.W.; Baek, M.; Yoon, S.H.; Park, K.H.; Lee, P.; et al. Efficient production of retinol in Yarrowia lipolytica by increasing stability using antioxidant and detergent extraction. Metab. Eng. 2022, 73, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.Y.; Zhang, T.L.; Yu, H.W. Selective biosynthesis of retinol in S. cerevisiae. Bioresour. Bioprocess. 2022, 22, 9–22. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, P.; Liu, L. Modular optimization of multi-gene pathways for fumarate production. Metab. Eng. 2016, 33, 76–85. [Google Scholar] [CrossRef]
- Nijland, J.G.; Shin, H.Y.; Jong, R.M.; Waal, P.P.; Klaassen, P.; Driessen, A.J. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol. Biofuels 2014, 7, 168. [Google Scholar] [CrossRef] [PubMed]
- Yoshikuni, Y.; Ferrin, T.E.; Keasling, J.D. Designed divergent evolution of enzyme function. Nature 2006, 440, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Tippmann, S.; Nielsen, J.; Khoomrung, S. Improved quantification of farnesene during microbial production from Saccharomyces cerevisiae in two liquid phase fermentations. Talanta 2016, 146, 100–106. [Google Scholar] [CrossRef]
- Jin, K.; Shi, X.; Liu, J.H.; Yu, W.W.; Liu, Y.F.; Li, J.H.; Du, G.C.; Lv, X.Q.; Liu, L. Combinatorial metabolic engineering enables the efficient production of ursolic acid and oleanolic acid in Saccharomyces cerevisiae. Bioresour. Technol. 2023, 374, 128819. [Google Scholar] [CrossRef]
- Rosenfeld, T.; Alchalal, A.; Ottolenghi, M. Primary photoprocesses in retinol. Chem. Phys. Lett. 1973, 20, 291–297. [Google Scholar] [CrossRef]
- Aksnes, L. Simultaneous determination of retinol, alpha-tocopherol, and 25-hydroxyvitamin D in human serum by high-performance liquid chromatography. J. Pediatr. Gastroenterol. Nutr. 1994, 18, 339–343. [Google Scholar] [CrossRef]
- Bu, X.; Lin, J.Y.; Cheng, J.; Yang, D.; Duan, C.Q.; Koffas, M.; Yan, G.L. Engineering endogenous ABC transporter with improving ATP supply and membrane flexibility enhances the secretion of β-carotene in Saccharomyces cerevisiae. Biotechnol. Biofuels 2020, 13, 168. [Google Scholar] [CrossRef]
- Tominaga, M.; Miyazaki, K.; Hataya, S.; Mitsui, Y.; Kuroda, S.; Kondo, A.; Ishii, J. Enhanced squalene production by modulation of pathways consuming squalene and its precursor. J. Biosci. Bioeng. 2022, 134, 1–6. [Google Scholar] [CrossRef] [PubMed]
- West, R.W.; Yocum, R.R.; Ptashne, M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: Location and function of the upstream activating sequence UASG. Mol. Cell. Biol. 1984, 4, 2467–2478. [Google Scholar] [PubMed]
- Torchia, T.E.; Hamilton, R.W.; Cano, C.L.; Hopper, J.E. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: Effects on carbon-controlled regulation of the galactose melibiose pathway genes. Mol. Cell. Biol. 1984, 4, 15–21. [Google Scholar]
- Chen, Y.; Xia, W.; Wang, Y.; Liu, H.; Li, X. Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb. Cell. Fact. 2016, 15, 113–121. [Google Scholar] [CrossRef]
- Kennedy, M.A.; Barbuch, R.; Bard, M. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1999, 1445, 110–122. [Google Scholar] [CrossRef]
- Salama, S.R.; Hendricks, K.B.; Thorner, J. G1 cyclin degradation: The PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell. Biol. 1994, 14, 53–66. [Google Scholar]
- Strand, M.K.; Stuart, G.R.; Longley, M.J.; Graziewicz, M.A.; Dominick, O.C.; Copeland, W.C. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot. Cell 2003, 2, 9–20. [Google Scholar] [CrossRef]
- Beis, K. Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 2015, 43, 89–93. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, X.; Zhang, C.; Zhou, X.; Xu, X.; Han, L.; Lv, X.; Liu, Y.; Liu, S.; Li, J.; et al. De novo biosynthesis of rubusoside and rebaudiosides in engineered yeasts. Nat. Commun. 2022, 13, 30–40. [Google Scholar] [CrossRef]
- Ikonomidis, A.; Tsakris, A.; Kanellopoulou, M.; Maniatis, A.N.; Pournaras, S. Effect of the proton motive force inhibitor carbonyl cyanide-m-chlorophenylhydrazone (CCCP) on Pseudomonas aeruginosa biofilm development. Lett. Appl. Microbiol. 2008, 47, 298–302. [Google Scholar] [CrossRef]
- Weidner, L.D.; Fung, K.L.; Kannan, P.; Moen, J.K.; Kumar, J.S.; Mulder, J.; Innis, R.B.; Gottesman, M.M.; Hall, M.D. Tariquidar is an inhibitor and not a substrate of human and mouse P-glycoprotein. Drug Metab. Dispos. 2016, 44, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Leus, I.V.; Weeks, J.W.; Bonifay, V.; Smith, L.; Richardson, S.; Zgurskaya, H.I. Substrate specificities and efflux efficiencies of RND efflux pumps of acinetobacter baumannii. J. Bacteriol. 2018, 200, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Ha, B.K.; Zhou, J.; Ahn, J.; Yoon, S.H.; Kim, S.W. Selective retinol production by modulating the composition of retinoids from metabolically engineered E. coli . Biotechnol. Bioeng. 2015, 112, 4–12. [Google Scholar]
- Zhao, Y.; Zhang, Y.; Nielsen, J.; Liu, Z. Production of β-carotene in Saccharomyces cerevisiae through altering yeast lipid metabolism. Biotechnol. Bioeng. 2021, 118, 2043–2052. [Google Scholar] [CrossRef]
- Bluemel, T.S.; Sanderson, M.Q.; Jensen, J.R.; Garrett, A.R.; Alegre, M.M.; Robison, R.A.; O’Neill, K.L. Abstract 4613: Serum levels of antioxidants in college aged, elderly, and cancer patients. Cancer Res. 2011, 71, 4603–4613. [Google Scholar] [CrossRef]
- Eisele, T.A.; Sinnhuber, R.O.; Nixon, J.E. Dietary antioxidant effects on the hepatic mixed-function oxidase system of rainbow trout. Food Chem. Toxicol. 1983, 21, 273–277. [Google Scholar] [CrossRef]
- Caramia, G.; Gori, A.; Valli, E.; Cerretani, L. Virgin olive oil in preventive medicine: From legend to epigenetics. Eur. J. Lipid Sci. Technol. 2012, 114, 375–388. [Google Scholar] [CrossRef]






| Strains | Characteristics | Reference |
|---|---|---|
| Escherichia coli JM109 | recA1, endA1, thi, gyrA96, supE44, hsdR17△ (lac-proAB)/F [traD36, proAB+, laclq, lacZ△M15] | Lab stock |
| S. cerevisiae CEN.PK2-1C | MATa, his3D1, leu2-3_112, ura3-52, trp1-289, MAL2-8c, SUC2 | Lab stock |
| S6 | S. cerevisiae CEN.PK2-1C derivate, ∆911b:: PTEF1-IDI, ∆PINO2:: PPGK-PTEF1-ERG20-PGPD-tHMG1, ∆ROX1:: PGPD-tHMG1 | Jin et al. [17] |
| A01 | S6 derivate, ∆308a:: PGAL7-crtE, ∆607c:: PGAL7-crtB, ∆GAL80:: PGAL7-crtI, ∆1309a:: PGAL7-crtYB, ∆1014a:: PGAL7-blh | This study |
| A02 | A01 derivate, ∆1021b:: PTEF1-ADH2 | This study |
| A03 | A02 derivate, ∆HIS3b:: PTEF1-ALD6 | This study |
| A04 | A03 derivate, ∆805a:: PTEF1-ACS1 | This study |
| A05 | A04 derivate, ∆CIT2 | This study |
| A06 | A05 derivate, ∆MLS1 | This study |
| A07 | A06 derivate, ∆CIT2 ∆MLS1 | This study |
| A08 | A07 derivate, ∆YPL062W | This study |
| A09 | A08 derivate, PERG9:: PHXT1 | This study |
| A10 | A08 derivate, PERG9:: PERG1 | This study |
| A11 | A08 derivate, PERG9:: PERG1-ERG9-CLN2 | This study |
| A12 | A11 derivate, ∆1414a:: PGAL7-crtE | This study |
| A13 | A11 derivate, ∆1414a:: crtE-PGAL1,10-crtI | This study |
| A14 | A13 derivate, ∆720a:: crtE-PGAL1,10-crtI | This study |
| A15 | A14 derivate, ∆1114a:: PGAL7-crtYB | This study |
| A16 | A15 derivate, ∆416d:: crtE-PGAL1,10-crtI | This study |
| A17 | A16 derivate, ∆106a:: PGAL7-crtYB | This study |
| A18 | A16 derivate, ∆1206a:: PGAL7-blh | This study |
| A19 | A16 derivate, ∆1206a:: blh-PGAL1,10-blh | This study |
| A20 | A19 derivate, ∆CAN1y:: PGAL7-POS5 | This study |
| AT1 | A20 derivate, ∆Pdr5 | This study |
| AT2 | A20 derivate, ∆Snq2 | This study |
| AT3 | A20 derivate, ∆Adp1 | This study |
| AT4 | A20 derivate, ∆Pdr10 | This study |
| AT5 | A20 derivate, ∆Mdl1 | This study |
| AT6 | A20 derivate, ∆Ste6 | This study |
| AT7 | A20 derivate, ∆Yor1 | This study |
| A21 | AT1 derivate, ∆1622b:: PGAL7-CYC2 | This study |
| A22 | AT1 derivate, ∆1622b:: PGAL7-FOX2 | This study |
| A23 | AT1 derivate, ∆1622b:: PGAL7-ENV9 | This study |
| A24 | AT1 derivate, ∆1622b:: PGAL7-IFA38 | This study |
| A25 | AT1 derivate, ∆1622b:: PGAL7-ybbo | This study |
| A26 | AT1 derivate, ∆1622b:: PGAL7-ENV9 (T99A), ∆208a:: PGAL7-ybbo | This study |
| A27 | A26 derivate, :: PTEF1-PAH1 | This study |
| A28 | A27 derivate, :: PTEF1-LRO1 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xu, X.; Liu, J.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Metabolic Engineering of Saccharomyces cerevisiae for Efficient Retinol Synthesis. J. Fungi 2023, 9, 512. https://doi.org/10.3390/jof9050512
Wang X, Xu X, Liu J, Liu Y, Li J, Du G, Lv X, Liu L. Metabolic Engineering of Saccharomyces cerevisiae for Efficient Retinol Synthesis. Journal of Fungi. 2023; 9(5):512. https://doi.org/10.3390/jof9050512
Chicago/Turabian StyleWang, Xuan, Xianhao Xu, Jiaheng Liu, Yanfeng Liu, Jianghua Li, Guocheng Du, Xueqin Lv, and Long Liu. 2023. "Metabolic Engineering of Saccharomyces cerevisiae for Efficient Retinol Synthesis" Journal of Fungi 9, no. 5: 512. https://doi.org/10.3390/jof9050512