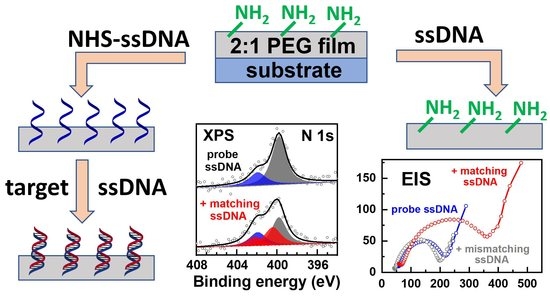
Rational Design of Porous Poly(ethylene glycol) Films as a Matrix for ssDNA Immobilization and Hybridization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Film Fabrication
2.3. ssDNA Immobilization and Hybridization
2.4. X-ray Photoelectron Spectroscopy
2.5. Electrochemistry
3. Results and Discussions
3.1. XPS
3.2. Electrochemical Studies

4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rao, A.N.; Grainger, D.W. Biophysical properties of nucleic acids at surfaces relevant to microarray performance. Biomater. Sci. 2014, 2, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, H. DNA Molecules site-specific immobilization and their applications. Cent. Eur. J. Chem. 2014, 12, 977–993. [Google Scholar] [CrossRef]
- Du, Y.; Dong, S. Nucleic acid biosensors: Recent advances and perspectives. Anal. Chem. 2017, 89, 189–215. [Google Scholar] [CrossRef] [PubMed]
- Herne, T.M.; Tarlov, M.J. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc. 1997, 119, 8916–8920. [Google Scholar] [CrossRef]
- Ballav, N.; Koelsch, P.; Zharnikov, M. Orientation and ordering in monomolecular films of sulfur-modified homo-oligonucleotides on gold. J. Phys. Chem. C 2009, 113, 18312–18320. [Google Scholar] [CrossRef]
- Schreiner, S.M.; Hatch, A.L.; Shudy, D.F.; Howard, D.R.; Howell, C.; Zhao, J.; Koelsch, P.; Zharnikov, M.; Petrovykh, D.Y.; Opdahl, A. Impact of DNA-surface interactions on the stability of DNA hybrids. Anal. Chem. 2011, 83, 4288–4295. [Google Scholar] [CrossRef]
- Leung, K.K.; Martens, I.; Yu, H.Z.; Bizzotto, D. Measuring and controlling the local environment of surface-bound DNA in self-assembled monolayers on gold when prepared using potential-assisted deposition. Langmuir 2020, 36, 6837–6847. [Google Scholar] [CrossRef]
- Umek, R.M.; Lin, S.W.; Vielmetter, J.; Terbrueggen, R.H.; Irvine, B.; Yu, C.J.; Kayyem, J.F.; Yowanto, H.; Blackburn, G.F.; Farkas, D.H.; et al. Electronic detection of nucleic acids. J. Mol. Diagn. 2001, 3, 74–84. [Google Scholar] [CrossRef]
- Raymundo-Pereira, P.A.; Oliveira Pedro, R.; Carr, O.; Melendez, M.E.; Gobbi, A.L.; Oliveira Piazzetta, M.H.; Oliveira, O.N., Jr. Influence of the molecular orientation and ionization of self-assembled monolayers in biosensors: Application to genosensors of prostate cancer antigen 3. J. Phys. Chem. C 2020, 125, 498–506. [Google Scholar] [CrossRef]
- Broderick, A.H.; Carter, M.C.D.; Lockett, M.R.; Smith, L.M.; Lynn, D.M. Fabrication of oligonucleotide and protein arrays on rigid and flexible substrates coated with reactive polymer multilayers. ACS Appl. Mater. Interfaces 2013, 5, 351–359. [Google Scholar] [CrossRef]
- Kargl, R.; Vorraber, V.; Ribitsch, V.; Köstler, S.; Stana-Kleinschek, K.; Mohan, T. Selective immobilization and detection of DNA on biopolymer supports for the design of microarrays. Biosens. Bioelectron. 2015, 68, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Cattani-Scholz, A.; Pedone, D.; Blobner, F.; Abstreiter, G.; Schwartz, J.; Tornow, M.; Andruzzi, L. PNA-PEG modified silicon platforms as functional bio-interfaces for applications in DNA microarrays and biosensors. Biomacromolecules 2009, 10, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sinsinbar, G.; Choudhary, M.; Kumar, V.; Pasricha, R.; Verma, H.N.; Arora, K. Graphene oxide-chitosan nanocomposite based electrochemical DNA biosensor for detection of typhoid. Sens. Actuators B 2013, 185, 675–684. [Google Scholar] [CrossRef]
- Park, S.R.; Hauver, J.; Zhang, Y.; Revyakin, A.; Coleman, R.A.; Tjian, R.; Chu, S.; Pertsinidis, A. A Single-molecule surface-based platform to detect the assembly and function of the human RNA polymerase II transcription machinery. Structure 2020, 28, 1337–1343. [Google Scholar] [CrossRef]
- Jeon, S.I.; Lee, J.H.; Andrade, J.D.; De Gennes, P.G. Protein-surface interactions in the presence of polyethylene oxide: I. Simplified theory. J. Colloid Interface Sci. 1991, 142, 149–158. [Google Scholar] [CrossRef]
- Polyethylene Glycol Chemistry: Biotechnical and Biomedical Applications. Harris, J.M. (Ed.) Plenum Press: New York, NY, USA, 1992; pp. 1–406. [Google Scholar]
- Holmberg, K.; Bergström, K.; Brink, C.; Österberg, E.; Tiberg, F.; Harris, J.M. Effects on protein adsorption, bacterial adhesion and contact angle of grafting PEG chains to polystyrene. J. Adhes. Sci. Technol. 1993, 7, 503–517. [Google Scholar] [CrossRef]
- Lowe, S.; O’Brien-Simpson, N.M.; Connal, L.A. Antibiofouling polymer interfaces: Poly(ethylene glycol) and other promising candidates. Polym. Chem. 2015, 6, 198–212. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Yu, B.; Wang, S.; Shen, Y.; Cong, H. Review of the research on anti-protein fouling coatings materials. Prog. Org. Coat. 2020, 147, 105860. [Google Scholar] [CrossRef]
- Prime, K.L.; Whitesides, G.M. Self-assembled organic monolayers: Model systems for studying adsorption of proteins at surfaces. Science 1991, 252, 1164–1167. [Google Scholar] [CrossRef]
- Harder, P.; Grunze, M.; Dahint, R.; Whitesides, G.M.; Laibinis, P.E. Molecular conformation in oligo(ethylene glycol)-terminated self-assembled monolayers on gold and silver surfaces determines their ability to resist protein adsorption. J. Phys. Chem. B 1998, 102, 426–436. [Google Scholar] [CrossRef]
- Herrwerth, S.; Eck, W.; Reinhardt, S.; Grunze, M. Factors that determine the protein resistance of oligoether self-assembled monolayers —internal hydrophilicity, terminal hydrophilicity, and lateral packing density. J. Am. Chem. Soc. 2003, 125, 9359–9366. [Google Scholar] [CrossRef]
- Schilp, S.; Rosenhahn, A.; Pettitt, M.E.; Bowen, J.; Callow, M.E.; Callow, J.A.; Grunze, M. Physicochemical properties of (ethylene glycol)-containing self-assembled monolayers relevant for protein and algal cell resistance. Langmuir 2009, 25, 10077–10082. [Google Scholar] [CrossRef]
- Rosenhahn, A.; Schilp, S.; Kreuzer, H.J.; Grunze, M. The Role of ‘‘inert’’ surface chemistry in marine biofouling prevention. Phys. Chem. Chem. Phys. 2010, 12, 4275–4286. [Google Scholar] [CrossRef]
- Khan, M.N.; Tjong, V.; Chilkoti, A.; Zharnikov, M. Fabrication of ssDNA/oligo(ethylene glycol) monolayers and complex nanostructures by an irradiation-promoted exchange reaction. Angew. Chem. Int. Ed. 2012, 51, 10303–10306. [Google Scholar] [CrossRef]
- Hu, W.-P.; Huang, L.-Y.; Kuo, T.-C.; Hu, W.-W.; Chang, Y.; Chen, C.-S.; Chen, H.-C.; Chen, W.-Y. Optimization of DNA-directed immobilization on mixed oligo(ethylene glycol) monolayers for immunodetection. Anal. Biochem. 2012, 423, 26–35. [Google Scholar] [CrossRef]
- Nagasaki, Y. Construction of a densely poly(ethylene glycol)-chain-tethered surface and its performance. Polymer J. 2011, 43, 949–958. [Google Scholar] [CrossRef]
- Jia, F.; Lu, X.; Tan, X.; Wang, D.; Cao, X.; Zhang, K. Effect of PEG architecture on the hybridization thermodynamics and protein accessibility of PEGylated oligonucleotides. Angew. Chem. Int. Ed. 2017, 56, 1239–1243. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, K. PEGylation of therapeutic oligonucletides: From linear to highly branched PEG architectures. Nano Res. 2018, 11, 5519–5534. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Jia, F.; Miller, A.; Tan, X.; Chen, P.; Zhang, L.; Lu, H.; Fang, Y.; Kang, X.; et al. Self-assembled DNA−PEG bottlebrushes enhance antisense activity and pharmacokinetics of oligonucleotides. ACS Appl. Mater. Interfaces 2020, 12, 45830–45837. [Google Scholar] [CrossRef]
- Kivlehan, F.; Paolucci, M.; Brennan, D.; Ragoussis, I.; Galvin, P. Three-dimensional hydrogel structures as optical sensor arrays, for the detection of specific DNA sequences. Anal. Biochem. 2012, 421, 1–8. [Google Scholar] [CrossRef]
- Wang, K.W.; Betancourt, T.; Hall, C.K. Computational study of DNA-cross-linked hydrogel formation for drug delivery applications. Macromolecules 2018, 51, 9758–9768. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, H.; Luan, C.; Fu, F.; Chen, B.; Liu, H.; Zhao, Y. Porous hydrogel encapsulated photonic barcodes for multiplex microRNA quantification. Adv. Funct. Mater. 2018, 28, 1704458. [Google Scholar] [CrossRef]
- Qi, Y.; Li, K.; Zhao, C.; Ma, Y.; Yang, W. Preparation of a poly (PEGDA-co-GMA) thin hydrogel matrix for oligonucleotide microarray applications. J. Chem. Technol. Biotechnol. 2021, 96, 1902–1908. [Google Scholar] [CrossRef]
- Meyerbröker, N.; Kriesche, T.; Zharnikov, M. Novel ultrathin polyethylene glycol films as flexible platform for biological applications and plasmonics. ACS Appl. Mater. Interfaces 2013, 5, 2641–2649. [Google Scholar] [CrossRef]
- Zhao, Z.; Das, S.; Zharnikov, M. Tuning the properties of polyethylene glycol films and membranes by molecular weight of the precursors. ACS Appl. Polymer Mater. 2021, 4, 645–653. [Google Scholar] [CrossRef]
- Meyerbröker, N.; Zharnikov, M. Modification and patterning of nanometer-thin poly(ethylene glycol) films by electron irradiation. ACS Appl. Mater. Interfaces 2013, 5, 5129–5138. [Google Scholar] [CrossRef] [PubMed]
- Meyerbröker, N.; Zharnikov, M. Ultraflexible, Freestanding nanomembranes based on poly(ethylene glycol). Adv. Mater. 2014, 26, 3328–3332. [Google Scholar] [CrossRef]
- Meyerbröker, N.; Zharnikov, M. Hydrogel nanomembranes as templates for patterned deposition of nanoparticles on arbitrary substrates. ACS Appl. Mater. Interfaces 2014, 6, 14729–14735. [Google Scholar] [CrossRef]
- Khan, M.; Schuster, S.; Zharnikov, M. Giant electrical response of pristine and nanoparticle-loaded hydrogel nanomembranes to humidity variation. J. Phys. Chem. C 2015, 119, 14427–14433. [Google Scholar] [CrossRef]
- Khan, M.; Schuster, S.; Zharnikov, M. Chemical derivatization and biofunctionalization of hydrogel nanomembranes for potential biomedical and biosensor applications. Phys. Chem. Chem. Phys. 2016, 18, 12035–12042. [Google Scholar] [CrossRef]
- Zhao, Z.; Yan, R.; Zharnikov, M. The effect of ultraviolet light on biorepulsive hydrogel poly(ethylene glycol) films. ACS Appl. Polymer Mater. 2021, 3, 3446–3454. [Google Scholar] [CrossRef]
- Mädler, S.; Bich, C.; Touboul, D.; Zenobi, R. Chemical cross-linking with NHS esters: A systematic study on amino acid reactivities. J. Mass Spectrom. 2009, 44, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zharnikov, M. Elastic properties of poly(ethylene glycol) nanomembranes and respective implications. Membranes 2022, 12, 509. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Gong, P.; Harbers, G.M.; Grainger, D.W.; Castner, D.G.; Gamble, L. Surface coverage and structure of mixed DNA/alkylthiol monolayers on gold: Characterization by XPS, NEXAFS, and fluorescence intensity measurements. J. Anal. Chem. 2006, 78, 3316–3325. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Nguyen, P.-C.T.; Grainger, D.W.; Gamble, L.J.; Castner, D.G. Structure and DNA hybridization properties of mixed nucleic acid/maleimide−ethylene glycol monolayers. Anal. Chem. 2007, 79, 4390–4400. [Google Scholar] [CrossRef]
- Schreiner, S.M.; Shudy, D.F.; Hatch, A.L.; Opdahl, A.; Whitman, L.J.; Petrovykh, D.Y. Controlled and efficient hybridization achieved with DNA probes immobilized solely through preferential DNA-substrate interactions. Anal. Chem. 2010, 82, 2803–2810. [Google Scholar] [CrossRef]
- Howell, C.; Zhao, J.; Koelsch, P.; Zharnikov, M. Hybridization in ssDNA films—a multi-technique spectroscopy study. Phys. Chem. Chem. Phys. 2011, 13, 15512–15522. [Google Scholar] [CrossRef]
- Moulder, J.F.; Stickle, W.E.; Sobol, P.E.; Bomben, K.D. Handbook of X-ray Photoelectron Spectroscopy. Chastian, J., Ed.; Perkin-Elmer-Corporation: Eden Prairie, MN, USA, 1992; pp. 184–185. [Google Scholar]
- Ossowski, J.; Wächter, T.; Silies, L.; Kind, M.; Noworolska, A.; Blobner, F.; Gnatek, D.; Rysz, J.; Bolte, M.; Feulner, P.; et al. Thiolate versus selenolate: Structure, stability and charge transfer properties. ACS Nano 2015, 9, 4508–4526. [Google Scholar] [CrossRef]
- Mohammadian, N.; Farnoush, F. ALS Genosensing using DNA-hybridization electrochemical biosensor based on label-free immobilization of ssDNA on Sm2O3 NPs-rGO/PANI composite. Sensor. Actuat. B Chem. 2018, 275, 432–438. [Google Scholar] [CrossRef]
- Saprigin, A.; Thomas, C.; Dulcey, C.; Patterson, C.; Spector, M. Spectroscopic quantification of covalently immobilized oligonucleotides. Surf. Interface Anal. 2004, 36, 24–32. [Google Scholar] [CrossRef]
- Petrovykh, D.Y.; Kimura-Suda, H.; Tarlov, M.J.; Whitman, L.J. Quantitative characterization of DNA films by X-ray photoelectron spectroscopy. Langmuir 2004, 20, 429–440. [Google Scholar] [CrossRef]
- Rei Vilar, M.; Botelho do Rego, A.M.; Ferraria, A.M.; Jugnet, Y.; Nogues, C.; Peled, D.; Naaman, R.J. Interaction of self-assembled monolayers of DNA with electrons: HREELS and XPS studies. J. Phys. Chem. B 2008, 112, 6957–6964. [Google Scholar] [CrossRef]
- Petrovykh, D.Y.; Perez-Dieste, V.; Opdahl, A.; Kimura-Suda, H.; Sullivan, J.M.; Tarlov, M.J.; Himpsel, F.J.; Whitman, L. Nucleobase orientation and ordering in films of single-stranded DNA on gold. J. Am. Chem. Soc. 2006, 128, 2–3. [Google Scholar] [CrossRef]
- Opdahl, A.; Petrovykh, D.Y.; Kimura-Suda, H.; Tarlov, M.J.; Whitman, L.J. Independent control of grafting density and conformation of single-stranded DNA brushes. Proc. Natl. Acad. Sci. USA. 2007, 104, 9–14. [Google Scholar] [CrossRef]
- Howell, C.; Jeyachandran, Y.L.; Koelsch, P.; Zharnikov, M. Orientation and Ordering in Sequence- and Length-Mismatched Surface-Bound DNA Hybrids. J. Phys. Chem. C 2012, 116, 11133–11140. [Google Scholar] [CrossRef]
- Howell, C.; Hamoudi, H.; Zharnikov, M. Thymine/adenine diblock-oligonucleotide monolayers and hybrid brushes on gold: A spectroscopic study. Biointerphases 2013, 8, 6. [Google Scholar] [CrossRef]
- Roth, E.; Glick Azaria, A.; Girshevitz, O.; Bitler, A.; Garini, Y. Measuring the conformation and persistence length of single-stranded DNA using a DNA origami structure. Nano Lett. 2018, 18, 6703–6709. [Google Scholar] [CrossRef]
- Petrovykh, D.Y.; Kimura-Suda, H.; Whitman, L.J.; Tarlov, M.J. Quantitative analysis and characterization of DNA immobilized on gold. J. Am. Chem. Soc. 2003, 125, 5219–5226. [Google Scholar] [CrossRef]
- Zhang, S.; Pan, N. Supercapacitors performance evaluation. Adv. En. Mater. 2015, 5, 1401401. [Google Scholar] [CrossRef]
- Liu, R.; Pan, S.; Liu, M.; Huang, W.; Lv, Z.; He, A. A Label-free electrochemical biosensor with magnetically induced self-assembly for the detection of CYP2C9* 3 gene. Appl. Surf. Sci. 2021, 537, 147868. [Google Scholar] [CrossRef]
- Irving, D.; Gong, P.; Levicky, R. DNA surface hybridization: Comparison of theory and experiment. J. Phys. Chem. B 2010, 114, 7631–7640. [Google Scholar] [CrossRef] [PubMed]
- Wong, I.Y.; Melosh, N.A. An electrostatic model for DNA surface hybridization. Biophys. J. 2010, 98, 2954–2963. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, T.J.; Knotts IV, T.A. Thermodynamics of DNA hybridization on surfaces. J. Chem. Phys. 2011, 134, 205105. [Google Scholar] [CrossRef] [PubMed]
- Freitas, R.R.Q.; Rivelino, R.; de Mota, F.B.; Gueorguiev, G.K.; de Castilho, C.M.C. Energy barrier reduction for the double proton-transfer reaction in guanine-cytosine DNA base pair on a gold surface. J. Phys. Chem. C 2015, 119, 15735–15741. [Google Scholar] [CrossRef]








| Sample | Relative Capacitance | Charge Transfer Resistance |
|---|---|---|
| Au | 100% | 25 Ω |
| Au/PEG | 67% | 41 Ω |
| Au/PEG+A10 | 65% | 40 Ω |
| Au/PEG+T10 | 65% | 43 Ω |
| Au/PEG/NHS-T5 | 50% | 95 Ω |
| Au/PEG/NHS-T5+T5 | 49% | 97 Ω |
| Au/PEG/NHS-T5+A5 | 32% | 195 Ω |
| Au/PEG/NHS-T10 | 34% | 158 Ω |
| Au/PEG/NHS-T10+T10 | 33% | 155 Ω |
| Au/PEG/NHS-T10+A10 | 19% | 330 Ω |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Das, S.; Zharnikov, M. Rational Design of Porous Poly(ethylene glycol) Films as a Matrix for ssDNA Immobilization and Hybridization. Bioengineering 2022, 9, 414. https://doi.org/10.3390/bioengineering9090414
Zhao Z, Das S, Zharnikov M. Rational Design of Porous Poly(ethylene glycol) Films as a Matrix for ssDNA Immobilization and Hybridization. Bioengineering. 2022; 9(9):414. https://doi.org/10.3390/bioengineering9090414
Chicago/Turabian StyleZhao, Zhiyong, Saunak Das, and Michael Zharnikov. 2022. "Rational Design of Porous Poly(ethylene glycol) Films as a Matrix for ssDNA Immobilization and Hybridization" Bioengineering 9, no. 9: 414. https://doi.org/10.3390/bioengineering9090414