Topology Optimization-Based Localized Bone Microstructure Reconstruction for Image Resolution Enhancement: Accuracy and Efficiency
Abstract
:1. Introduction
2. Materials and Methods
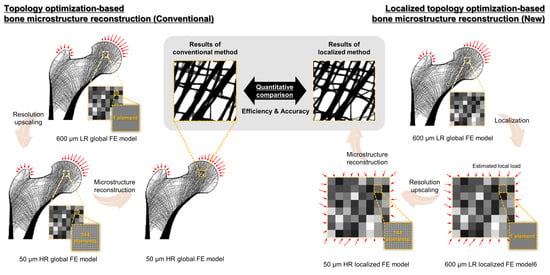
2.1. Conventional Topology Optimization-Based Bone Microstructure Reconstruction Using the Global Model
2.2. Novel Topology Optimization-Based Localized Bone Microstructure Reconstruction Using the Localized Model
2.3. Numerical Validation Based on Quantitative Comparison Using Proximal Femur
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ammann, P.; Rizzoli, R. Bone Strength and Its Determinants. Osteoporos. Int. 2003, 14, 13–18. [Google Scholar] [CrossRef]
- Goldstein, S.A.; Goulet, R.; McCubbrey, D. Measurement and Significance of Three-Dimensional Architecture to the Mechanical Integrity of Trabecular Bone. Calcif. Tissue Int. 1993, 53, S127–S133. [Google Scholar] [CrossRef] [Green Version]
- Goulet, R.W.; Goldstein, S.A.; Ciarelli, M.J.; Kuhn, J.L.; Brown, M.B.; Feldkamp, L.A. The Relationship between the Structural and Orthogonal Compressive Properties of Trabecular Bone. J. Biomech. 1994, 27, 375–389. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.W.; Kim, J.J. Structural Behavior Analysis According to Porous Structures of the Bone Scaffold in the Femoral Head. J. Korean Soc. Precis. Eng. 2022, 39, 627–633. [Google Scholar] [CrossRef]
- Weinans, H.H. Mechanically Induced Bone Adaptations around Orthopaedic Implants. Ph.D. Thesis, University of Nijmegen, Nijmegen, The Netherlands, 1991. [Google Scholar]
- Peyrin, F.; Salome, M.; Cloetens, P.; Laval-Jeantet, A.M.; Ritman, E.; Rüegsegger, P. Micro-CT Examinations of Trabecular Bone Samples at Different Resolutions: 14, 7 and 2 Micron Level. Technol. Health Care 1998, 6, 391–401. [Google Scholar] [CrossRef]
- Bouxsein, M.L.; Boyd, S.K.; Christiansen, B.A.; Guldberg, R.E.; Jepsen, K.J.; Müller, R. Guidelines for Assessment of Bone Microstructure in Rodents Using Micro-Computed Tomography. J. Bone Miner. Res. 2010, 25, 1468–1486. [Google Scholar] [CrossRef] [PubMed]
- Krug, R.; Burghardt, A.J.; Majumdar, S.; Link, T.M. High-Resolution Imaging Techniques for the Assessment of Osteoporosis. Radiol. Clin. N. Am. 2010, 48, 601–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Xu, L.; Chen, E. Image Denoising and Inpainting with Deep Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 1, 341–349. [Google Scholar]
- Clark, J.L.; Wadhwani, C.P.; Abramovitch, K.; Rice, D.D.; Kattadiyil, M.T. Effect of Image Sharpening on Radiographic Image Quality. J. Prosthet. Dent. 2018, 120, 927–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Ren, J.S.J.; Liu, C.; Jia, J. Deep Convolutional Neural Network for Image Deconvolution. Adv. Neural Inf. Process. Syst. 2014, 2, 1790–1798. [Google Scholar]
- Agarwal, M.; Mahajan, R. Medical Image Contrast Enhancement Using Range Limited Weighted Histogram Equalization. Procedia Comput. Sci. 2018, 125, 149–156. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Weng, W.; Zhu, X. INet: Convolutional Networks for Biomedical Image Segmentation. IEEE Access 2021, 9, 16591–16603. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. ChemInform 2015, 37, 1951–1954. [Google Scholar] [CrossRef]
- Umehara, K.; Ota, J.; Ishida, T. Application of Super-Resolution Convolutional Neural Network for Enhancing Image Resolution in Chest CT. J. Digit. Imaging 2018, 31, 441–450. [Google Scholar] [CrossRef]
- Mozaffari, S.; Al-Jarrah, O.Y.; Dianati, M.; Jennings, P.; Mouzakitis, A. Deep Learning-Based Vehicle Behavior Prediction for Autonomous Driving Applications: A Review. IEEE Trans. Intell. Transp. Syst. 2022, 23, 33–47. [Google Scholar] [CrossRef]
- Pritt, M.; Chern, G. Satellite Image Classification with Deep Learning. In Proceedings of the 2017 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 10–12 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–7. [Google Scholar]
- Abdolahnejad, M.; Liu, P.X. Deep Learning for Face Image Synthesis and Semantic Manipulations: A Review and Future Perspectives. Artif. Intell. Rev. 2020, 53, 5847–5880. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [Green Version]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep Learning for Medical Image Processing: Overview, Challenges and the Future. In Classification in BioApps; Springer: Cham, Switzerland, 2018; Volume 26, pp. 323–350. ISBN 9783319659817. [Google Scholar]
- Moran, M.; Faria, M.; Giraldi, G.; Bastos, L.; Conci, A. Do Radiographic Assessments of Periodontal Bone Loss Improve with Deep Learning Methods for Enhanced Image Resolution? Sensors 2021, 21, 2013. [Google Scholar] [CrossRef]
- Hildebrand, T.; Laib, A.; Müller, R.; Dequeker, J.; Rüegsegger, P. Direct Three-Dimensional Morphometric Analysis of Human Cancellous Bone: Microstructural Data from Spine, Femur, Iliac Crest, and Calcaneus. J. Bone Miner. Res. 1999, 14, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Verhulp, E.; van Rietbergen, B.; Huiskes, R. Load Distribution in the Healthy and Osteoporotic Human Proximal Femur during a Fall to the Side. Bone 2008, 42, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Bagi, C.M.; Berryman, E.; Moalli, M.R. Comparative Bone Anatomy of Commonly Used Laboratory Animals: Implications for Drug Discovery. Comp. Med. 2011, 61, 76–85. [Google Scholar] [PubMed]
- Levchuk, A.; Zwahlen, A.; Weigt, C.; Lambers, F.M.; Badilatti, S.D.; Schulte, F.A.; Kuhn, G.; Müller, R. The Clinical Biomechanics Award 2012—Presented by the European Society of Biomechanics: Large Scale Simulations of Trabecular Bone Adaptation to Loading and Treatment. Clin. Biomech. 2014, 29, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Jang, I.G. Image Resolution Enhancement for Healthy Weight-Bearing Bones Based on Topology Optimization. J. Biomech. 2016, 49, 3035–3040. [Google Scholar] [CrossRef]
- Wolff, J. Das Gesetz Der Transformation Der Knochen. DMW—Dtsch. Med. Wochenschr. 1893, 19, 1222–1224. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.J.; Nam, J.; Jang, I.G. Computational Study of Estimating 3D Trabecular Bone Microstructure for the Volume of Interest from CT Scan Data. Int. J. Numer. Methods Biomed. Eng. 2018, 34, e2950. [Google Scholar] [CrossRef]
- Kim, J.J.; Kim, Y.; Jang, I.G. Estimation of Local Bone Loads for the Volume of Interest. J. Biomech. Eng. 2016, 138, 071004. [Google Scholar] [CrossRef]
- Verhulp, E.; van Rietbergen, B.; Huiskes, R. Comparison of Micro-Level and Continuum-Level Voxel Models of the Proximal Femur. J. Biomech. 2006, 39, 2951–2957. [Google Scholar] [CrossRef]
- Bendsøe, M.P. Optimization of Structural Topology, Shape, and Material; Springer: Berlin/Heidelberg, Germany, 1995; ISBN 978-3-662-03117-9. [Google Scholar]
- Park, J.; Sutradhar, A.; Shah, J.J.; Paulino, G.H. Design of Complex Bone Internal Structure Using Topology Optimization with Perimeter Control. Comput. Biol. Med. 2018, 94, 74–84. [Google Scholar] [CrossRef]
- Yoon, S.; Schiffer, A.; Kim, J.J.; Jang, I.G.; Lee, S.; Kim, T.-Y. Numerical Predictions of the Interaction between Highly Nonlinear Solitary Waves and the Microstructure of Trabecular Bone in the Femoral Head. J. Mech. Behav. Biomed. Mater. 2020, 109, 103805. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, Y.; Kim, J.J.; Jang, I.G. Homeostasis-Based Aging Model for Trabecular Changes and Its Correlation with Age-Matched Bone Mineral Densities and Radiographs. Eur. J. Radiol. 2015, 84, 2261–2268. [Google Scholar] [CrossRef]
- Yi, C.; Wang, M.; Wei, J.; Wang, J.; Wang, L.; Cheng, X. Preoperative QCT Assessment of Femoral Head for Assessment of Femoral Head Bone Loss. Exp. Ther. Med. 2017, 13, 1470–1474. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Lee, I.; Ha, S.H.; Park, J.H.; Park, J.H. Comparison of Hip Subregion Bone Mineral Density to the Type of Proximal Femur Fracture. Arch. Osteoporos. 2020, 15, 122. [Google Scholar] [CrossRef]
- Chen, Y.; Pani, M.; Taddei, F.; Mazzà, C.; Li, X.; Viceconti, M. Large-Scale Finite Element Analysis of Human Cancellous Bone Tissue Micro Computer Tomography Data: A Convergence Study. J. Biomech. Eng. 2014, 136, 101013. [Google Scholar] [CrossRef]
- Beaupré, G.S.; Orr, T.E.; Carter, D.R. An Approach for Time-Dependent Bone Modeling and Remodeling-Theoretical Development. J. Orthop. Res. 1990, 8, 651–661. [Google Scholar] [CrossRef]
- Tsubota, K.; Adachi, T.; Tomita, Y. Functional Adaptation of Cancellous Bone in Human Proximal Femur Predicted by Trabecular Surface Remodeling Simulation toward Uniform Stress State. J. Biomech. 2002, 35, 1541–1551. [Google Scholar] [CrossRef]
- Hestenes, M.R.; Stiefel, E. Methods of Conjugate Gradients for Solving Linear Systems. J. Res. Natl. Bur. Stand. 1952, 49, 409. [Google Scholar] [CrossRef]
- Svanberg, K. The Method of Moving Asymptotes—A New Method for Structural Optimization. Int. J. Numer. Methods Eng. 1987, 24, 359–373. [Google Scholar] [CrossRef]
- Armstrong, C.G. Modelling Requirements for Finite-Element Analysis. Comput. Des. 1994, 26, 573–578. [Google Scholar] [CrossRef]
- Jang, I.G.; Kim, I.Y.; Kwak, B.M. Analogy of Strain Energy Density Based Bone-Remodeling Algorithm and Structural Topology Optimization. J. Biomech. Eng. 2009, 131, 011012. [Google Scholar] [CrossRef]
- Jang, I.G.; Kim, I.Y. Computational Simulation of Simultaneous Cortical and Trabecular Bone Change in Human Proximal Femur during Bone Remodeling. J. Biomech. 2010, 43, 294–301. [Google Scholar] [CrossRef]





| Region of Interest | Index | 4.8 × 4.8 mm2 | 9.6 × 9.6 mm2 | 14.4 × 14.4 mm2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Conv. 1 Result | Loc. 2 Result | Imp. 3 (%) | Conv. 1 Result | Loc. 2 Result | Imp. 3 (%) | Conv. 1 Result | Loc. 2 Result | Imp. 3 (%) | ||
| Femoral head | Time (h) | 22.8 | 2.0 | 91.23 | 21.6 | 1.3 | 93.98 | 22.8 | 2.7 | 88.16 |
| Iteration | 363 | 689 | - | 320 | 457 | - | 351 | 607 | - | |
| Resource (GB) | 56.4 | 0.3 | 99.47 | 56.1 | 1.0 | 98.22 | 56.3 | 2.3 | 95.91 | |
| Femoral neck | Time (h) | 13.3 | 0.7 | 94.74 | 13.2 | 0.8 | 93.94 | 21.7 | 1.8 | 91.71 |
| Iteration | 200 | 233 | - | 210 | 289 | - | 278 | 474 | - | |
| Resource (GB) | 56.4 | 0.3 | 99.47 | 56.1 | 1.0 | 98.22 | 56.3 | 2.3 | 95.91 | |
| Intertrochanter | Time (h) | 22.9 | 1.8 | 92.14 | 21.1 | 1.1 | 94.79 | 27.6 | 1.4 | 94.93 |
| Iteration | 317 | 648 | - | 343 | 362 | - | 372 | 353 | - | |
| Resource (GB) | 56.4 | 0.3 | 99.47 | 56.1 | 1.0 | 98.22 | 56.3 | 2.3 | 95.91 | |
| Region of Interest | Index | 4.8 × 4.8 mm2 | 9.6 × 9.6 mm2 | 14.4 × 14.4 mm2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Conv. 1 Result | Loc. 2 Result | Error (%) | Conv. 1 Result | Loc. 2 Result | Error (%) | Conv. 1 Result | Loc. 2 Result | Error (%) | ||
| Femoral head | BV/TV (%) | 56.89 | 56.87 | 0.02 | 56.41 | 56.37 | 0.04 | 54.96 | 54.93 | 0.03 |
| Tb.Th (μm) | 262.65 | 245.34 | 6.59 | 268.84 | 255.73 | 4.88 | 249.06 | 241.76 | 2.93 | |
| Tb.Sp (μm) | 312.65 | 292.30 | 6.51 | 326.25 | 310.87 | 4.71 | 320.63 | 311.54 | 2.84 | |
| Tb.N (mm−1) | 2.17 | 2.32 | 6.91 | 2.10 | 2.20 | 4.76 | 2.21 | 2.27 | 2.71 | |
| Femoral neck | BV/TV (%) | 21.14 | 20.86 | 0.28 | 20.02 | 19.93 | 0.09 | 22.30 | 22.21 | 0.09 |
| Tb.Th (μm) | 113.97 | 95.81 | 15.93 | 125.18 | 110.21 | 11.96 | 141.04 | 132.16 | 6.30 | |
| Tb.Sp (μm) | 667.81 | 571.04 | 14.49 | 785.63 | 695.62 | 11.46 | 772.05 | 727.30 | 5.80 | |
| Tb.N (mm−1) | 1.85 | 2.18 | 17.84 | 1.60 | 1.81 | 13.12 | 1.58 | 1.68 | 6.33 | |
| Intertrochanter | BV/TV (%) | 50.31 | 50.25 | 0.06 | 40.59 | 40.53 | 0.06 | 35.40 | 35.36 | 0.04 |
| Tb.Th (μm) | 192.65 | 181.59 | 5.74 | 169.00 | 158.39 | 6.28 | 165.57 | 155.71 | 5.96 | |
| Tb.Sp (μm) | 298.85 | 282.43 | 5.49 | 388.62 | 365.00 | 6.08 | 474.70 | 447.04 | 5.83 | |
| Tb.N (mm−1) | 2.61 | 2.77 | 6.13 | 2.40 | 2.56 | 6.67 | 2.14 | 2.27 | 6.07 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Kim, J.J. Topology Optimization-Based Localized Bone Microstructure Reconstruction for Image Resolution Enhancement: Accuracy and Efficiency. Bioengineering 2022, 9, 644. https://doi.org/10.3390/bioengineering9110644
Kim J, Kim JJ. Topology Optimization-Based Localized Bone Microstructure Reconstruction for Image Resolution Enhancement: Accuracy and Efficiency. Bioengineering. 2022; 9(11):644. https://doi.org/10.3390/bioengineering9110644
Chicago/Turabian StyleKim, Jisun, and Jung Jin Kim. 2022. "Topology Optimization-Based Localized Bone Microstructure Reconstruction for Image Resolution Enhancement: Accuracy and Efficiency" Bioengineering 9, no. 11: 644. https://doi.org/10.3390/bioengineering9110644