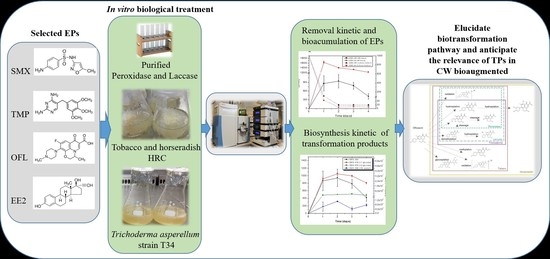
Investigating the Transformation Products of Selected Antibiotics and 17 α-Ethinylestradiol under Three In Vitro Biotransformation Models for Anticipating Their Relevance in Bioaugmented Constructed Wetlands
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Biological Material
2.3. Emerging Pollutant Removal by HRC, T. asperellum, and Pure Enzymes
2.4. Extraction and Analysis of Emerging Pollutants and Their Transformation Products
2.5. Determination of Plant POD and Fungal Laccase Activity
2.6. Confirmation of Glycosylated Transformation Products and Indirect Quantification
2.7. Data Analysis
3. Results and Discussion
3.1. EP Removal from the Medium and EP Bioaccumulation in Tissues
3.2. Analysis of Transformation Products (TPs) Produced by HRC and T. asperellum
3.3. Confirmation of Glucose Conjugated-TPs and Indirect Quantification by Beta-Glucosidase Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bavumiragira, J.P.; Ge, J.; Yin, H. Fate and transport of pharmaceuticals in water systems: A processes review. Sci. Total Environ. 2022, 823, 153635. [Google Scholar] [CrossRef] [PubMed]
- Gulde, R.; Rutsch, M.; Clerc, B.; Schollée, J.E.; von Gunten, U.; McArdell, C.S. Formation of transformation products during ozonation of secondary wastewater effluent and their fate in post-treatment: From laboratory-to full-scale. Water Res. 2021, 200, 117200. [Google Scholar] [CrossRef] [PubMed]
- Manasfi, T.; Houska, J.; Gebhardt, I.; von Gunten, U. Formation of carbonyl compounds during ozonation of lake water and wastewater: Development of a non-target screening method and quantification of target compounds. Water Res. 2023, 237, 119751. [Google Scholar] [CrossRef]
- Polińska, W.; Kotowska, U.; Kiejza, D.; Karpińska, J. Insights into the Use of Phytoremediation Processes for the Removal of Organic Micropollutants from Water and Wastewater; A Review. Water 2021, 13, 2065. [Google Scholar] [CrossRef]
- Carvalho, P.N. Constructed Wetlands and Phytoremediation as a Tool for Pharmaceutical Removal. In Hanbook of Environmental Chemistry; Springer: Berlin/Heidelberg, Germany, 2020; Volume 103, pp. 317–413. [Google Scholar] [CrossRef]
- Zhao, X.; Bai, S.; Li, C.; Yang, J.; Ma, F. Bioaugmentation of atrazine removal in constructed wetland: Performance, microbial dynamics, and environmental impacts. Bioresour. Technol. 2019, 289, 121618. [Google Scholar] [CrossRef]
- Tondera, K.; Chazarenc, F.; Chagnon, P.-L.; Brisson, J. Bioaugmentation of treatment wetlands—A review. Sci. Total Environ. 2021, 775, 145820. [Google Scholar] [CrossRef]
- Ávila, C.; García-Galán, M.J.; Uggetti, E.; Montemurro, N.; García-Vara, M.; Pérez, S.; García, J.; Postigo, C. Boosting pharmaceutical removal through aeration in constructed wetlands. J. Hazard. Mater. 2021, 412, 125231. [Google Scholar] [CrossRef]
- Tadić, Đ.; Sauvêtre, A.; Cerqueira, F.; Lestremau, F.; Ait-Mouheb, N.; Chiron, S. Partially saturated vertical surface flow con-structed wetland for emerging contaminants and antibiotic resistance genes removal from wastewater: The effect of bioaug-mentation with Trichoderma fungus. Ecol. Eng. 2023; submitted. [Google Scholar]
- Morsi, R.; Bilal, M.; Iqbal, H.M.; Ashraf, S.S. Laccases and peroxidases: The smart, greener and futuristic biocatalytic tools to mitigate recalcitrant emerging pollutants. Sci. Total Environ. 2020, 714, 136572. [Google Scholar] [CrossRef]
- Manasfi, R.; Chiron, S.; Montemurro, N.; Perez, S.; Brienza, M. Biodegradation of fluoroquinolone antibiotics and the climbazole fungicide by Trichoderma species. Environ. Sci. Pollut. Res. 2020, 27, 23331–23341. [Google Scholar] [CrossRef]
- Piyaviriyakul, P.; Boontanon, N.; Boontanon, S.K. Bioremoval and tolerance study of sulfamethoxazole using whole cell Trichoderma harzianum isolated from rotten tree bark. J. Environ. Sci. Health Part A 2021, 56, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Sauvêtre, A.; May, R.; Harpaintner, R.; Poschenrieder, C.; Schröder, P. Metabolism of carbamazepine in plant roots and endophytic rhizobacteria isolated from Phragmites australis. J. Hazard. Mater. 2018, 342, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Antony, S.; Antony, S.; Rebello, S.; George, S.; Biju, D.T.; Reshmy, R.; Madhavan, A.; Binod, P.; Pandey, A.; Sindhu, R.; et al. Bioremediation of Endocrine Disrupting Chemicals—Advancements and Challenges. Environ. Res. 2022, 213, 113509. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma spp. for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Alsayed, A.; Weisskopf, L. Deciphering trichoderma–plant–pathogen interactions for better development of biocontrol apremoval through aeration in constructed wetlands. J. Fungi. 2021, 7, 61. [Google Scholar] [CrossRef]
- Alderete, L.G.S.; Talano, M.A.; Ibáñez, S.G.; Purro, S.; Agostini, E.; Milrad, S.R.; Medina, M.I. Establishment of transgenic tobacco hairy roots expressing basic peroxidases and its application for phenol removal. J. Biotechnol. 2009, 139, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Balcázar-López, E.; Méndez-Lorenzo, L.H.; Batista-García, R.A.; Esquivel-Naranjo, U.; Ayala, M.; Kumar, V.V.; Savary, O.; Cabana, H.; Herrera-Estrella, A.; Folch-Mallol, J.L. Xenobiotic Compounds Degradation by Heterologous Expression of a Trametes sanguineus Laccase in Trichoderma atroviride. PLoS ONE 2016, 11, e0147997. [Google Scholar] [CrossRef] [Green Version]
- Campos, L.; López-Gresa, M.P.; Fuertes, D.; Bellés, J.M.; Rodrigo, I.; Lisón, P. Tomato glycosyltransferase Twi1 plays a role in flavonoid glycosylation and defence against virus. BMC Plant Biol. 2019, 19, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Huang, H.; Chen, C.-E.; Qi, S.; Price, O.R.; Zhang, H.; Jones, K.C.; Sweetman, A.J. Simultaneous determination of 20 trace organic chemicals in waters by solid-phase extraction (SPE) with triple-quadrupole mass spectrometer (QqQ-MS) and hybrid quadrupole Orbitrap high resolution MS (Q-Orbitrap-HRMS). Chemosphere 2016, 163, 99–107. [Google Scholar] [CrossRef]
- Hurtado, C.; Domínguez, C.; Clapés, P.; Bayona, J.M. Determination of the β-glycosylate fraction of contaminants of emerging concern in lettuce (Lactuca sativa L.) grown under controlled conditions. Anal. Bioanal. Chem. 2018, 410, 5715–5721. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.; Reinhold, D. Metabolism of Sulfamethoxazole by the Model Plant Arabidopsis thaliana. Environ. Sci. Technol. 2019, 53, 4901–4911. [Google Scholar] [CrossRef]
- Dudley, S.; Sun, C.; Jiang, J.; Gan, J. Metabolism of sulfamethoxazole in Arabidopsis thaliana cells and cucumber seedlings. Environ. Pollut. 2018, 242, 1748–1757. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Acharya, K. Algae-mediated removal of selected pharmaceutical and personal care products (PPCPs) from Lake Mead water. Sci. Total. Environ. 2017, 581–582, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Kiki, C.; Rashid, A.; Wang, Y.; Li, Y.; Zeng, Q.; Yu, C.-P.; Sun, Q. Dissipation of antibiotics by microalgae: Kinetics, identification of transformation products and pathways. J. Hazard. Mater. 2020, 387, 121985. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Pandey, B.; Suthar, S. Phytotoxicity and degradation of antibiotic ofloxacin in duckweed (Spirodela polyrhiza) system. Ecotoxicol. Environ. Saf. 2019, 179, 88–95. [Google Scholar] [CrossRef]
- Wang, P.; Wong, Y.-S.; Tam, N.F.-Y. Green microalgae in removal and biotransformation of estradiol and ethinylestradiol. J. Appl. Phycol. 2017, 29, 263–273. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Q.; Li, Y.; Wang, H.; Wu, K.; Yu, C.-P. Biotransformation of estrone, 17β-estradiol and 17α-ethynylestradiol by four species of microalgae. Ecotoxicol. Environ. Saf. 2019, 180, 723–732. [Google Scholar] [CrossRef]
- Castellana, G.; Loffredo, E. Simultaneous Removal of Endocrine Disruptors from a Wastewater Using White Rot Fungi and Various Adsorbents. Water Air Soil Pollut. 2014, 225, 1872. [Google Scholar] [CrossRef]
- Lv, Y.; Li, Y.; Liu, X.; Xu, K. The tolerance mechanism and accumulation characteristics of Phragmites australis to sulfamethoxazole and ofloxacin. Chemosphere 2020, 253, 126695. [Google Scholar] [CrossRef]
- Sauvêtre, A.; Eichhorn, P.; Pérez, S. Metabolism of Pharmaceuticals in Plants and Their Associated Microbiota. In Interaction and Fate of Pharmaceuticals in Soil-Crop Systems; The Handbook of Environmental Chemistry; Pérez Solsona, S., Montemurro, N., Chiron, S., Barceló, D., Eds.; Springer: Cham, Switzerland, 2020; Volume 103. [Google Scholar] [CrossRef]
- Spina, F.; Gea, M.; Bicchi, C.; Cordero, C.; Schilirò, T.; Varese, G.C. Ecofriendly laccases treatment to challenge micropollutants issue in municipal wastewaters. Environ. Pollut. 2020, 257, 113579. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Preis, M.; Harvey, P.J.; Grosse, S.; Letzel, T.; Schröder, P. Emerging pollutants and plants—Metabolic activation of diclofenac by peroxidases. Chemosphere 2016, 146, 435–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zdarta, J.; Jankowska, K.; Strybel, U.; Marczak, Ł.; Nguyen, L.N.; Oleskowicz-Popiel, P.; Jesionowski, T. Bioremoval of estrogens by laccase immobilized onto polyacrylonitrile/polyethersulfone material: Effect of inhibitors and mediators, process characterization and catalytic pathways determination. J. Hazard. Mater. 2022, 432, 128688. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, J. Kinetics and mechanisms for sulfamethoxazole transformation in the phenolic acid-laccase (Trametes versicolor) system. Environ. Sci. Pollut. Res. 2022, 29, 62941–62951. [Google Scholar] [CrossRef] [PubMed]
- Tadić, Đ.; Gramblicka, M.; Mistrik, R.; Bayona, J.M. Systematic identification of trimethoprim metabolites in lettuce. Anal. Bioanal. Chem. 2022, 414, 3121–3135. [Google Scholar] [CrossRef]
- Tian, R.; Zhang, R.; Uddin, M.; Qiao, X.; Chen, J.; Gu, G. Uptake and metabolism of clarithromycin and sulfadiazine in lettuce. Environ. Pollut. 2019, 247, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Klampfl, C.W. Metabolization of pharmaceuticals by plants after uptake from water and soil: A review. Trends Anal. Chem. 2019, 111, 13–26. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Sandermann, H., Jr. Higher plant metabolism of xenobiotics: The ‘green liver’ concept. Pharmacogenetics 1994, 4, 225–241. [Google Scholar] [CrossRef]
- Boiteau, R.M.; Hoyt, D.W.; Nicora, C.D.; Kinmonth-Schultz, H.A.; Ward, J.K.; Bingol, K. Structure Elucidation of Unknown Metabolites in Metabolomics by Combined NMR and MS/MS Prediction. Metabolites 2018, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Albreht, A.; Hussain, H.; Jiménez, B.; Yuen, A.H.Y.; Whiley, L.; Witt, M.; Lewis, M.R.; Chekmeneva, E. Structure Elucidation and Mitigation of Endogenous Interferences in LC-MS-Based Metabolic Profiling of Urine. Anal. Chem. 2022, 94, 1760–1768. [Google Scholar] [CrossRef]
- Maculewicz, J.; Kowalska, D.; Świacka, K.; Toński, M.; Stepnowski, P.; Białk-Bielińska, A.; Dołżonek, J. Transformation products of pharmaceuticals in the environment: Their fate, (eco)toxicity and bioaccumulation potential. Sci. Total Environ. 2022, 802, 149916. [Google Scholar] [CrossRef] [PubMed]
- Göbel, A.; Thomsen, A.; McArdell, C.S.; Joss, A.; Giger, W. Occurrence and Sorption Behavior of Sulfonamides, Macrolides, and Trimethoprim in Activated Sludge Treatment. Environ. Sci. Technol. 2005, 39, 3981–3989. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Ye, Q.; Zhang, J.; Richards, J.; Borchardt, D.; Gan, J. Diclofenac in Arabidopsis cells: Rapid formation of conjugates. Environ. Pollut. 2017, 222, 383–392. [Google Scholar] [CrossRef]
- Reis, A.C.; Kolvenbach, B.A.; Nunes, O.C.; Corvini, P.F. Biodegradation of antibiotics: The new resistance determinants—Part II. New Biotechnol. 2020, 54, 13–27. [Google Scholar] [CrossRef]
- Maury, S.; Delaunay, A.; Mesnard, F.; Crônier, D.; Chabbert, B.; Geoffroy, P.; Legrand, M. O-methyltransferase(s)-suppressed plants produce lower amounts of phenolic vir inducers and are less susceptible to Agrobacterium tumefaciens infection. Planta 2010, 232, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Ezelarab, H.; Abbas, S.; Hassan, H.A.; Abuo-Rahma, G.E.-D.A. Recent updates of fluoroquinolones as antibacterial agents. Arch. Pharm. 2018, 351, e1800141. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alderete, L.S.; Sauvêtre, A.; Chiron, S.; Tadić, Đ. Investigating the Transformation Products of Selected Antibiotics and 17 α-Ethinylestradiol under Three In Vitro Biotransformation Models for Anticipating Their Relevance in Bioaugmented Constructed Wetlands. Toxics 2023, 11, 508. https://doi.org/10.3390/toxics11060508
Alderete LS, Sauvêtre A, Chiron S, Tadić Đ. Investigating the Transformation Products of Selected Antibiotics and 17 α-Ethinylestradiol under Three In Vitro Biotransformation Models for Anticipating Their Relevance in Bioaugmented Constructed Wetlands. Toxics. 2023; 11(6):508. https://doi.org/10.3390/toxics11060508
Chicago/Turabian StyleAlderete, Lucas Sosa, Andrés Sauvêtre, Serge Chiron, and Đorđe Tadić. 2023. "Investigating the Transformation Products of Selected Antibiotics and 17 α-Ethinylestradiol under Three In Vitro Biotransformation Models for Anticipating Their Relevance in Bioaugmented Constructed Wetlands" Toxics 11, no. 6: 508. https://doi.org/10.3390/toxics11060508