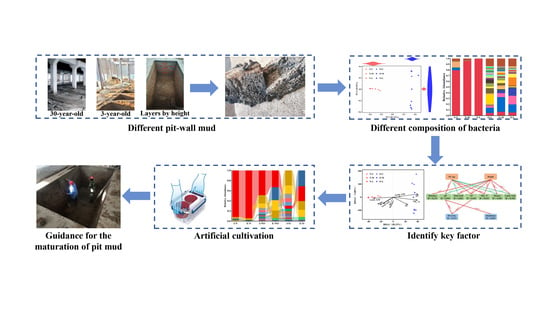
Bacterial Communities Found in Pit-Wall Mud and Factors Driving Their Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. PWM Samples
2.2. Illumina MiSeq Sequencing
2.3. Assessment of Physicochemical Properties
2.4. Statistical Analysis
2.5. Driving the Evolution of Bacterial Communities in PWM
2.6. Accession Numbers
3. Results
3.1. Diversity of Bacterial Communities in Different PWM Samples
3.2. Composition of Bacterial Communities in Different PWM Samples
3.3. Differences and Interactions between NPWM and OPWM
3.4. Physicochemical Properties of Different PWM Samples
3.5. Correlations between Bacterial Communities and Physicochemical Properties
3.6. Complex Correlations Observed in PWM
3.7. Evolution of Bacterial Communities in Different PWM Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, J.; Liu, G.; Li, A.; Liang, C.; Ren, C.; Xu, Y. Domination of pit mud microbes in the formation of diverse flavour compounds during Chinese strong aroma-type Baijiu fermentation. LWT—Food Sci. Technol. 2021, 137, 110442. [Google Scholar] [CrossRef]
- Xu, B.; Xu, S.; Cai, J.; Sun, W.; Mu, D.; Wu, X.; Li, X. Analysis of the microbial community and the metabolic profile in medium-temperature Daqu after inoculation with Bacillus licheniformis and Bacillus velezensis. LWT—Food Sci. Technol. 2022, 160, 113214. [Google Scholar] [CrossRef]
- Lu, M.; Zhou, W.; Ji, F.; Wu, J.; Nie, Y.; Ren, C.; Xu, Y. Profiling prokaryotic community in pit mud of Chinese strong-aroma type liquor by using oligotrophic culturing. Int. J. Food Microbiol. 2021, 337, 108951. [Google Scholar] [CrossRef]
- Wang, X.; Du, H.; Xu, Y. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int. J. Food Microbiol. 2017, 244, 27–35. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Z.; Wen, S.; Ouyang, G.; Shen, Y.; Yang, Q.; Ren, C.; Xu, Y. Rare short- and medium-chain fatty acid-producing anaerobes from raw soil play vital roles in formation of diverse flavour compounds of Jiangxiangxing Baijiu. Food Microbiol. 2023, 112, 104247. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, M.; Xu, B.; Liu, L.; Sun, W.; Mu, D.; Wu, X.; Li, X. Microbial communities and flavor formation in the fermentation of Chinese strong-flavor Baijiu produced from old and new Zaopei. Food Res. Int. 2022, 156, 111162. [Google Scholar] [CrossRef]
- Ding, X.; Wu, C.; Huang, J.; Zhou, R. Characterization of interphase volatile compounds in Chinese Luzhou-flavor liquor fermentation cellar analyzed by head space-solid phase micro extraction coupled with gas chromatography mass spectrometry (HS-SPME/GC/MS). LWT—Food Sci. Technol. 2016, 66, 124–133. [Google Scholar] [CrossRef]
- Gao, J.; Qin, J.; Ye, F.; Ding, F.; Liu, G.; Li, A.; Ren, C.; Xu, Y. Constructing simplified microbial consortia to improve the key flavour compounds during strong aroma-type Baijiu fermentation. Int. J. Food Microbiol. 2022, 369, 109594. [Google Scholar] [CrossRef]
- Tao, Y.; Li, J.; Rui, J.; Xu, Z.; Zhou, Y.; Hu, X.; Wang, X.; Liu, M.; Li, D.; Li, X.; et al. Prokaryotic Communities in Pit Mud from Different-Aged Cellars Used for the Production of Chinese Strong-Flavored Liquor. Appl. Environ. Microbiol. 2014, 80, 2254–2260. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Wang, X.; Li, X.; Wei, N.; Jin, H.; Xu, Z.; Tang, Q.; Zhu, X. The functional potential and active populations of the pit mud microbiome for the production of Chinese strong-flavour liquor. Microb. Biotechnol. 2017, 10, 1603–1615. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Meng, Y.; Wang, Y.; Zhou, Q.; Li, A.; Liu, G.; Li, J.; Xing, X. Prokaryotic communities in multidimensional bottom-pit-mud from old and young pits used for the production of Chinese Strong-Flavor Baijiu. Food Chem. 2020, 312, 126084. [Google Scholar] [CrossRef]
- Gao, Z.; Wu, Z.; Zhang, W. Effect of Pit Mud on Bacterial Community and Aroma Components in Yellow Water and Their Changes during the Fermentation of Chinese Strong-Flavor Liquor. Foods 2020, 9, 372. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wu, X.; Mu, D.; Yang, W.; Jiang, S.; Sun, W.; Shen, Y.; Cai, J.; Zheng, Z.; Jiang, S.; et al. Profiling the effects of physicochemical indexes on the microbial diversity and its aroma substances in pit mud. Lett. Appl. Microbiol. 2020, 71, 667–678. [Google Scholar] [CrossRef]
- Cai, W.; Xue, Y.; Tang, F.; Wang, Y.; Yang, S.; Liu, W.; Hou, Q.; Yang, X.; Guo, Z.; Shan, C. The Depth-Depended Fungal Diversity and Non-depth-Depended Aroma Profiles of Pit Mud for Strong-Flavor Baijiu. Front. Microbiol. 2022, 12, 789845. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Q.; Wang, Q.; Li, C.; Leng, Y.; Li, S.; Zhou, X.; Han, W.; Li, J.; Zhang, X.; et al. Long-term batch brewing accumulates adaptive microbes, which comprehensively produce more flavorful Chinese liquors. Food Res. Int. 2014, 62, 894–901. [Google Scholar] [CrossRef]
- Chai, L.; Qian, W.; Zhong, X.; Zhang, X.; Lu, Z.; Zhang, S.; Wang, S.; Shen, C.; Shi, J.; Xu, Z.; et al. Mining the Factors Driving the Evolution of the Pit Mud Microbiome under the Impact of Long-Term Production of Strong-Flavor Baijiu. Appl. Environ. Microbiol. 2021, 87, e00885-21. [Google Scholar] [CrossRef]
- Wu, Q.; Zhu, Y.; Fang, C.; Wijffels, R.; Xu, Y. Can we control microbiota in spontaneous food fermentation?—Chinese liquor as a case example. Trends Food Sci. Technol. 2021, 110, 321–331. [Google Scholar] [CrossRef]
- Zou, W.; Zhao, C.; Luo, H. Diversity and Function of Microbial Community in Chinese Strong-Flavor Baijiu Ecosystem: A Review. Front. Microbiol. 2018, 9, 671. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Tang, Y.; Guo, X.; Zhao, K.; Penttinen, P.; Tian, X.; Zhang, X.; Ren, D.; Zhang, X.; Ercolini, D. Structural and Functional Changes in Prokaryotic Communities in Artificial Pit Mud during Chinese Baijiu Production. mSystems 2020, 5, e00829-19. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Jin, Z.; Ali, A.; Wang, C.; Liu, J. Caproic Acid-Producing Bacteria in Chinese Baijiu Brewing. Front. Microbiol. 2022, 13, 883142. [Google Scholar] [CrossRef]
- Tan, G.; Zhou, R.; Zhang, W.; Hu, Y.; Ruan, Z.; Li, J.; Zhang, C.; Shen, D.; Peng, N.; Liang, Y.; et al. Detection of Viable and Total Bacterial Community in the Pit Mud of Chinese Strong-Flavor Liquor Using Propidium Monoazide Combined With Quantitative PCR and 16S rRNA Gene Sequencing. Front. Microbiol. 2020, 11, 896. [Google Scholar] [CrossRef]
- Zhu, X.; Zhou, Y.; Wang, Y.; Wu, T.; Li, X.; Li, D.; Tao, Y. Production of high-concentration n-caproic acid from lactate through fermentation using a newly isolated Ruminococcaceae bacterium CPB6. Biotechnol. Biofuels 2017, 10, 102. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, Z.; Yu, Z.; Shen, G.; Cheng, H.; Tao, S. Composition and diversity of soil microbial communities in the alpine wetland and alpine forest ecosystems on the Tibetan Plateau. Sci. Total Environ. 2020, 747, 141358. [Google Scholar] [CrossRef]
- Imhoff, J. New Dimensions in Microbial Ecology—Functional Genes in Studies to Unravel the Biodiversity and Role of Functional Microbial Groups in the Environment. Microorganisms 2016, 4, 19. [Google Scholar] [CrossRef]
- Hu, X.; Du, H.; Ren, C.; Xu, Y.; Björkroth, J. Illuminating Anaerobic Microbial Community and Cooccurrence Patterns across a Quality Gradient in Chinese Liquor Fermentation Pit Muds. Appl. Environ. Microbiol. 2016, 82, 2506–2515. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhao, H.; Xue, S.; Chen, K.; Zhang, Y. Effection of Lactic Acid Dissociation on Swelling-Based Short-Chain Fatty Acid Vesicles Nano-Delivery. Foods 2022, 11, 1630. [Google Scholar] [CrossRef]
- Poeplau, C.; Herrmann, A.; Kätterer, T. Opposing effects of nitrogen and phosphorus on soil microbial metabolism and the implications for soil carbon storage. Soil Biol. Biochem. 2016, 100, 83–91. [Google Scholar] [CrossRef]
- Wang, L.; Wen, Y.; Tong, R.; Zhang, H.; Chen, H.; Hu, T.; Liu, G.; Wang, J.; Zhu, L.; Wu, T. Understanding Responses of Soil Microbiome to the Nitrogen and Phosphorus Addition in Metasequoia glyptostroboides Plantations of Different Ages. Microb. Ecol. 2022, 84, 565–579. [Google Scholar] [CrossRef]
- Zou, W.; Ye, G.; Zhang, K. Diversity, Function, and Application of Clostridium in Chinese Strong Flavor Baijiu Ecosystem: A Review. J. Food Sci. 2018, 83, 1193–1199. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sun, M.; Hou, P.; Wang, W.; Shen, X.; Zhang, L.; Han, S.; Pan, C. Analysis of microbial community structure and volatile compounds in pit mud used for manufacturing Taorong-type Baijiu based on high-throughput sequencing. Sci. Rep. 2022, 12, 7347. [Google Scholar] [CrossRef]
- Hicks, L.; Frey, B.; Kjøller, R.; Lukac, M.; Moora, M.; Weedon, J.; Rousk, J. Toward a function-first framework to make soil microbial ecology predictive. Ecol. 2022, 103, e03594. [Google Scholar] [CrossRef]
- Wang, H.; Gu, Y.; Zhou, W.; Zhao, D.; Qiao, Z.; Zheng, J.; Gao, J.; Chen, X.; Ren, C.; Xu, Y.; et al. Adaptability of a Caproate-Producing Bacterium Contributes to Its Dominance in an Anaerobic Fermentation System. Appl. Environ. Microbiol. 2021, 87, e01203-21. [Google Scholar] [CrossRef]
- Ji, M.; Du, H.; Xu, Y. Structural and metabolic performance of p-cresol producing microbiota in different carbon sources. Food Res. Int. 2020, 132, 109049. [Google Scholar] [CrossRef]
- Pang, X.; Han, B.; Huang, X.; Zhang, X.; Hou, L.; Cao, M.; Gao, L.; Hu, G.; Chen, J. Effect of the environment microbiota on the flavour of light-flavour Baijiu during spontaneous fermentation. Sci. Rep. 2018, 8, 3396. [Google Scholar] [CrossRef] [Green Version]
- Pang, X.; Chen, C.; Huang, X.; Yan, Y.; Chen, J.; Han, B. Influence of indigenous lactic acid bacteria on the volatile flavor profile of light-flavor Baijiu. LWT—Food Sci. Technol. 2021, 147, 111540. [Google Scholar] [CrossRef]
- Koyanagi, T.; Nakagawa, A.; Kiyohara, M.; Matsui, H.; Tsuji, A.; Barla, F.; Take, H.; Katsuyama, Y.; Tokuda, K.; Nakamura, S.; et al. Tracing microbiota changes in yamahai-moto, the traditional Japanese sake starter. Biosci. Biotechnol. Biochem. 2016, 80, 399–406. [Google Scholar] [CrossRef]
- Zhou, W.; Xia, Y.; Zhao, Y.; Wang, Y.; Wu, Z.; Suyama, T.; Zhang, W. Study on the Effect of Key Genes ME2 and adhE during Luzhou-Flavor Baijiu Brewing. Foods 2022, 11, 700. [Google Scholar] [CrossRef]
- Avila, M.; Sanchez, C.; Calzada, J.; Mayer, M.; Berruga, M.; Lopez-Diaz, T.; Narbad, A.; Garde, S. Isolation and characterization of new bacteriophages active against Clostridium tyrobutyricum and their role in preventing the late blowing defect of cheese. Food Res. Int. 2023, 163, 112222. [Google Scholar] [CrossRef]
- Andersen, S.; De Groof, V.; Khor, W.; Roume, H.; Props, R.; Coma, M.; Rabaey, K. A Clostridium Group IV Species Dominates and Suppresses a Mixed Culture Fermentation by Tolerance to Medium Chain Fatty Acids Products. Front. Microbiol. 2017, 5, 8. [Google Scholar] [CrossRef] [Green Version]




| Sample | NPWM | OPWM | |||||
|---|---|---|---|---|---|---|---|
| Parameters | Upper | Middle | Lower | Upper | Middle | Lower | |
| Moisture (%) | 39.78 ± 0.88 b | 39.81 ± 2.18 b | 39.52 ± 0.25 b | 35.94 ± 1.80 c | 36.29 ± 0.77 c | 44.05 ± 0.98 a | |
| pH | 4.03 ± 0.05 d | 3.92 ± 0.05 e | 3.77 ± 0.00 f | 5.20 ± 0.00 a | 5.05 ± 0.06 b | 4.61 ± 0.06 c | |
| Total Acid (mg/g) | 10.10 ± 0.37 c | 14.60 ± 0.28 b | 20.50 ± 0.57 a | 7.50 ± 0.13 d | 7.92 ± 0.91 d | 19.76 ± 0.51 a | |
| AP (mg/100 g) | 241.07 ± 36.92 bc | 70.32 ± 9.74 d | 256.17 ± 58.25 bc | 196.37 ± 40.06 cd | 381.10 ± 62.01 b | 985.00 ± 113.87 a | |
| NH4+-N (mg/100 g) | 26.97 ± 2.88 c | 35.09 ± 6.41 c | 24.82 ± 1.56 c | 61.03 ± 2.36 b | 75.01 ± 1.44 a | 80.48 ± 8.35 a | |
| Humus (%) | 10.99 ± 0.85 b | 15.90 ± 0.55 a | 17.28 ± 1.29 a | 8.35 ± 0.66 c | 11.81 ± 0.58 b | 15.75 ± 0.60 a | |
| Lactic Acid (mg/g) | 9.43 ± 0.29 c | 11.65 ± 2.30 b | 20.56 ± 0.50 a | 6.44 ± 0.18 d | 2.93 ± 0.60 e | 8.68 ± 0.33 c | |
| Acetic Acid (mg/g) | 1.60 ± 0.51 b | 1.74 ± 0.08 b | 4.40 ± 0.33 a | 4.43 ± 0.42 a | 1.71 ± 0.27 b | 4.57 ± 0.95 a | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Xu, B.; Xu, S.; Jiang, S.; Mu, D.; Wu, X.; Li, X. Bacterial Communities Found in Pit-Wall Mud and Factors Driving Their Evolution. Foods 2023, 12, 1419. https://doi.org/10.3390/foods12071419
Zhou H, Xu B, Xu S, Jiang S, Mu D, Wu X, Li X. Bacterial Communities Found in Pit-Wall Mud and Factors Driving Their Evolution. Foods. 2023; 12(7):1419. https://doi.org/10.3390/foods12071419
Chicago/Turabian StyleZhou, Hao, Boyang Xu, Shanshan Xu, Suwei Jiang, Dongdong Mu, Xuefeng Wu, and Xingjiang Li. 2023. "Bacterial Communities Found in Pit-Wall Mud and Factors Driving Their Evolution" Foods 12, no. 7: 1419. https://doi.org/10.3390/foods12071419