Assignment of a Reference Value of Total Cow’s Milk Protein Content in Baked Cookies Used in an Interlaboratory Comparison
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Reference Solutions
2.3. Preparation of the Test Material
2.4. Preparation of Sample and Calibration Blends
2.5. Measurements
2.6. Computations and Statistical Analysis
2.7. Assigned Value (xa)
2.8. Associated Measurement Uncertainty of the Assigned Value (u(xa))
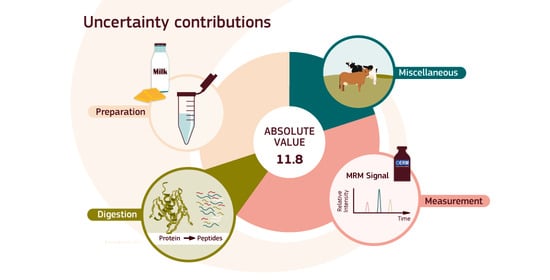
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crevel, R.W.R.; Ballmer-Weber, B.K.; Holzhauser, T.; Hourihane, J.O.; Knulst, A.C.; Mackie, A.; Timmermans, F.; Taylor, S.L. Thresholds for food allergens and their value to different stakeholders. Allergy 2008, 63, 597–609. [Google Scholar] [CrossRef]
- Remington, B.C.; Westerhout, J.; Meima, M.Y.; Blom, W.M.; Kruizinga, A.G.; Wheeler, M.W.; Taylor, S.L.; Houben, G.; Baumert, J.L. Updated population minimal eliciting dose distributions for use in risk assessment of 14 priority food allergens. Food Chem. Toxicol. 2020, 139, 111259. [Google Scholar] [CrossRef]
- International Organization for Standardization. ISO/IEC 17025—General Requirements for the Competence of Testing and Calibration Laboratories. 2017. Available online: https://www.iso.org/files/live/sites/isoorg/files/store/en/PUB100424.pdf (accessed on 16 March 2022).
- Owen, L.; Gilbert, J. Proficiency testing for quality assurance of allergens methods. Anal. Bioanal. Chem. 2009, 395, 147–153. [Google Scholar] [CrossRef]
- Cordeiro, F.; Cubero-Leon, E.; Nørgaard, J.; Martinez-Esteso, M.J.; Brohée, M.; Breidbach, A.; Cizek-Stroh, A.; O’Connor, G.; Robouch, P.; Emons, H. Total cow’s milk protein in cookies: The first interlaboratory comparison with a well-defined measurand fit for food allergen risk assessment. Accreditation Qual. Assur. 2021, 26, 177–181. [Google Scholar] [CrossRef]
- Grebe, S.K.; Singh, R.J. Clinical peptide and protein quantification by mass spectrometry (MS). TrAC Trends Anal. Chem. 2016, 84, 131–143. [Google Scholar] [CrossRef]
- Seger, C.; Salzmann, L. After another decade: LC–MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, A.N.; Cobbaert, C.M.; Delatour, V.; Kelleher, N.L.; Lowenthal, M.S.; Shuford, C.M. Should LC-MS/MS Be the Reference Measurement Procedure to Determine Protein Concentrations in Human Samples? Clin. Chem. 2020, 67, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Heumann, K.G. Isotope dilution mass spectrometry of inorganic and organic substances. Anal. Bioanal. Chem. 1986, 325, 661–666. [Google Scholar] [CrossRef]
- Mackay, L.G.; Taylor, C.P.; Myors, R.B.; Hearn, R.; King, B. High accuracy analysis by isotope dilution mass spectrometry using an iterative exact matching technique. Accredit. Qual. Assur. 2003, 8, 191–194. [Google Scholar] [CrossRef]
- Vogl, J.; Pritzkow, W. Isotope dilution mass spectrometry—A primary method of measurement and its role for RM certification. MAPAN 2010, 25, 135–164. [Google Scholar] [CrossRef]
- Milton, M.; Quinn, T.J. Primary methods for the measurement of amount of substance. Metrologia 2001, 38, 289–296. [Google Scholar] [CrossRef]
- Martinez-Esteso, M.J.; O’Connor, G.; Nørgaard, J.; Breidbach, A.; Brohée, M.; Cubero-Leon, E.; Nitride, C.; Robouch, P.; Emons, H. A reference method for determining the total allergenic protein content in a processed food: The case of milk in cookies as proof of concept. Anal. Bioanal. Chem. 2020, 412, 8249–8267. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, A.; Kral, R.; Schimmel, H. Quantification of protein calibrants by amino acid analysis using isotope dilution mass spectrometry. Anal. Biochem. 2010, 408, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, A.N.; Whiteaker, J.R.; Carr, S.A.; Kuhn, E.; Liu, T.; Massoni, S.A.; Thomas, S.; Townsend, R.R.; Zimmerman, L.J.; Boja, E.S.; et al. Recommendations for the Generation, Quantification, Storage, and Handling of Peptides Used for Mass Spectrometry—Based Assays. Clin. Chem. 2016, 62, 48–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, Z.; Hopley, C.; O’Connor, G. High accuracy determination of malachite green and leucomalachite green in salmon tissue by exact matching isotope dilution mass spectrometry. J. Chromatogr. B 2008, 874, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Henrion, A. Reduction of systematic errors in quantitative analysis by isotope dilution mass spectrometry (IDMS): An iterative method. Anal. Bioanal. Chem. 1994, 350, 657–658. [Google Scholar] [CrossRef]
- Nitride, C.; Nørgaard, J.; Omar, J.; Emons, H.; Esteso, M.-J.M.; O’Connor, G. An assessment of the impact of extraction and digestion protocols on multiplexed targeted protein quantification by mass spectrometry for egg and milk allergens. Anal. Bioanal. Chem. 2019, 411, 3463–3475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.R-project.org/ (accessed on 16 March 2022).
- International Organization for Standardization. ISO 13528—Statistical Methods for Use in Proficiency Testing by Interlaboratory Comparison. 2015. Available online: https://www.iso.org/standard/56125.html (accessed on 16 March 2022).
- Kragten, J. Tutorial review. Calculating standard deviations and confidence intervals with a universally applicable spreadsheet technique. Analyst 1994, 119, 2161–2165. [Google Scholar] [CrossRef]
- Ellison, S.L.R.; Williams, A. (Eds.) Eurachem/CITAC Guide: Quantifying Uncertainty in Analytical Measurement, 3rd ed. 2012. ISBN 978-0-948926-30-3. Available online: https://www.eurachem.org (accessed on 16 March 2022).
- Gellrich, K.; Meyer, H.; Wiedemann, S. Composition of major proteins in cow milk differing in mean protein concentration during the first 155 days of lactation and the influence of season as well as short-term restricted feeding in early and mid-lactation. Czech J. Anim. Sci. 2014, 59, 97–106. [Google Scholar] [CrossRef] [Green Version]
- International Organization for Standardization. ISO/IEC Guide 98-3:2008. Uncertainty of Measurement—Part 3: Guide to the Expression of Uncertainty in Measurement (GUM:1995). Available online: https://www.iso.org/standard/50461.html (accessed on 16 March 2022).



| Protein a | Peptide | Position b | ic | jd |
|---|---|---|---|---|
| αS1-casein (CASA1_BOVIN) | FFVAPFPEVFGK | 38–49 | 1 | 1 |
| YLGYLEQLLR | 106–115 | 2 | 1 | |
| αS2-casein (CASA2_BOVIN) | ALNEINQFYQR | 96–106 | 1 | 2 |
| NAVPITPTLNR | 130–140 | 2 | 2 | |
| FALPQYLK | 189–196 | 3 | 2 | |
| VIPYVR | 215–220 | 4 | 2 | |
| β-casein (CASB_BOVIN) | VLPVPQK | 185–191 | 1 | 3 |
| AVPYPQR | 192–198 | 2 | 3 | |
| κ-casein (CASK_BOVIN) | YIPIQYVLSR | 46–55 | 1 | 4 |
| β-lactoglobulin (LACB_BOVIN) | IPAVFK | 94–99 | 1 | 5 |
| ALPMHIR | 158–164 | 2 | 5 |
| Eq. | Term | x | u(x) | Unit | RSU | Index | |
|---|---|---|---|---|---|---|---|
| FFV in TCMP,6 | 1 | mY,6 | 2.495 × 10−1 | 1.00 × 10−4 | g | 0.04% | <1% |
| bZc2,1,1 | 3.293 × 10−11 | 4.19 × 10−13 | mol/g | 1.3% | 92% | ||
| mZc2 | 2.515 × 10−1 | 1.00 × 10−4 | g | 0.04% | <1% | ||
| mYc2 | 2.506 × 10−1 | 1.00 × 10−4 | g | 0.04% | <1% | ||
| bZc1,1,1 | 3.890 × 10−12 | 4.97 × 10−14 | mol/g | 1.3% | 1% | ||
| mZc1 | 2.495 × 10−1 | 1.00 × 10−4 | g | 0.04% | <1% | ||
| mYc1 | 2.512 × 10−1 | 1.00 × 10−4 | g | 0.04% | <1% | ||
| R′1,1,6 | 4.579 × 10−1 | 1.72 × 10−3 | 0.4% | 6% | |||
| CASA1 in TCMP,6 | 2 | n1,1,6 | 3.334 × 10−12 | 5.00 × 10−14 | mol | 1.5% | <1% |
| n2,1,6 | 4.236 × 10−12 | 7.74 × 10−13 | mol | 18% | 37% | ||
| N1 | 2 | ||||||
| mExsolv,6 | 1.492 × 101 | 1.30 × 10−4 | g | 0.00% | <1% | ||
| mX,6 | 1.032 | 1.30 × 10−4 | g | 0.01% | <1% | ||
| mExtr,6 | 2.613 × 10−1 | 1.00 × 10−4 | g | 0.04% | <1% | ||
| M1 | 2.080 × 104 | 1.25 × 103 | g/mol | 6.0% | 13% | ||
| fP-P,1,6 | 1 | 1.19 × 10−1 | 12% | 50% | |||
| 5 proteins in TCMP,6 | 3 | w1,6 | 4.356 × 10−6 | 7.32 × 10−7 | g/g | 17% | 53% |
| w2,6 | 5.925 × 10−7 | 4.88 × 10−8 | g/g | 8.2% | <1% | ||
| w3,6 | 3.755 × 10−6 | 3.19 × 10−7 | g/g | 8.5% | 10% | ||
| w4,6 | 1.743 × 10−6 | 1.80 × 10−7 | g/g | 10% | 3% | ||
| w5,6 | 6.769 × 10−7 | 2.66 × 10−7 | g/g | 39% | 7% | ||
| fε | 1.087 | 5.02 × 10−2 | 4.6% | 26% | |||
| 6 independent TCMP values | 4 | wTCMP,6 | 1.209 × 10−5 | 1.09 × 10−6 | g/g | 9% | 15% |
| wTCMP,5 | 1.187 × 10−5 | 1.04 × 10−6 | g/g | 9% | 13% | ||
| wTCMP,4 | 1.196 × 10−5 | 1.25 × 10−6 | g/g | 10% | 19% | ||
| wTCMP,3 | 1.222 × 10−5 | 1.22 × 10−6 | g/g | 10% | 19% | ||
| wTCMP,2 | 1.134 × 10−5 | 9.41 × 10−7 | g/g | 8% | 11% | ||
| wTCMP,1 | 1.120 × 10−5 | 8.88 × 10−7 | g/g | 8% | 10% | ||
| fSB-SB | 1 | 1.43 × 10−2 | 1.4% | 13% | |||
| Result | xa | 11.8 | 0.55 | mg/kg | 4.7% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Breidbach, A.; Nørgaard, J.V.; Cubero-Leon, E.; Martinez Esteso, M.J. Assignment of a Reference Value of Total Cow’s Milk Protein Content in Baked Cookies Used in an Interlaboratory Comparison. Foods 2022, 11, 869. https://doi.org/10.3390/foods11060869
Breidbach A, Nørgaard JV, Cubero-Leon E, Martinez Esteso MJ. Assignment of a Reference Value of Total Cow’s Milk Protein Content in Baked Cookies Used in an Interlaboratory Comparison. Foods. 2022; 11(6):869. https://doi.org/10.3390/foods11060869
Chicago/Turabian StyleBreidbach, Andreas, Jørgen Vinther Nørgaard, Elena Cubero-Leon, and Maria Jose Martinez Esteso. 2022. "Assignment of a Reference Value of Total Cow’s Milk Protein Content in Baked Cookies Used in an Interlaboratory Comparison" Foods 11, no. 6: 869. https://doi.org/10.3390/foods11060869