Support vector regression (SVR), as an extension of the support vector machines in regressions, is commonly applied in a variety of fields [
42]. BPNN, as a known machine learning method, is proven to have great potential in rapid detection of meat adulteration [
14,
20]. Therefore, an SVR model with RBF kernel and a BPNN model were also examined in this study. The performances of the SVR model, RFR model, BPNN model, and 1DCNN framework were compared with that of the proposed 1DCNN-RFR framework using multiple evaluation metrics, i.e.,
, RMSE, and MAE. The 1DCNN framework consisted of the 1DCNN backbone and a 1DCNN regressor. The 1DCNN regressor consisted of two fully-connected layers with 64 and 32 neurons, respectively. ReLU and Sigmoid were selected as the activation functions for the first and second fully connected layers to strengthen the nonlinear expression ability of the 1DCNN regressor. A grid search method was used for identifying the best parameters of the five models via a three-fold cross-validation using the training set. The five models were programmed using the scikit learn library and the open-source PyTorch framework. Two experiments (Experiment A and Experiment B) with two datasets (Dataset A and Dataset B) were conducted to thoroughly compare the performances of the five models.
4.2.1. Experiment A
Dataset A was used in Experiment A, and was divided into two parts, namely the training set (data from the first 7 days, 147 samples) and the test set (data from the remaining 3 days, 63 samples). For the SVR, RFR, and BPNN models, the training set was expressed as a 1470 × 10 matrix, the test set was expressed as a 630 × 10 matrix. For the 1DCNN and 1DCNN-RFR frameworks, the training set and the test set were obtained from all the response values and were expressed as a 147 × 10 × 100 matrix and a 63 × 10 × 100 matrix, respectively.
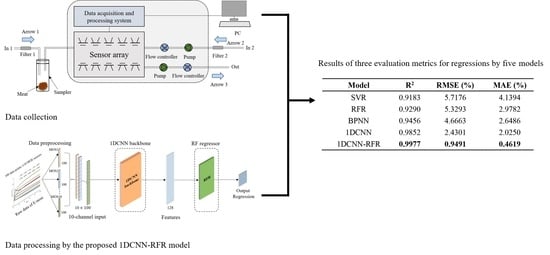
The SVR, RFR, and BPNN models, combined with SVs, were used to study the quantitative detection of adulteration in meat. The parameters searched for by the SVR model were the penalty factor (1, 5, 10, 20, 50, 100, 200, 500) and gamma (0.01, 0.1, 1, 5, 10, 20). The parameters searched for by the RFR model were the max depth (3, 5, 7, 9, 11, 13) and the minimum number of samples required to split an internal node (min. samples split: 7, 14, 21, 28, 35, 42). Through the grid search skill, the penalty factor and gamma for the SVR model were set to 50 and 1, respectively. For the RFR model, the max depth and the min samples split were set to 5 and 35, respectively. For the BPNN model, the optimum network architecture was obtained with topological architecture 10-21-1. An MSE loss function serves to calculate error and is minimized by the stochastic gradient descent (SGD) optimizer. ReLU was selected as the activation function of the BPNN model. The regression results from the SVR, RFR, and BPNN models on the test set are shown in
Table 5. Three evaluation metrics, the
, RMSE, and MAE, were computed to comprehensively and accurately assess these two regression models. When combined with SVs, the BPNN model achieved a marginally better result with an
of 0.9456, an RMSE of 4.6663%, and a MAE of 2.6486%. However, the performances of these three models were unsatisfactory because they failed to extract a sufficient number of features.
The 10-channel input was submitted to the proposed 1DCNN framework consisting of the 1DCNN backbone and a 1DCNN regressor. The 1DCNN framework was implemented on a laptop (Intel Core i7-9750H processor). The number of epochs was 1100 and the training time was 57.6207 s. The training loss of the 1DCNN framework during the training process is shown in
Figure 6. The 1DCNN framework converged after 920 epochs. The parameters at 920 epochs were saved and used to evaluate the performance of the 1DCNN framework on the test set. As shown in
Table 5, the 1DCNN framework performed much better than the SVR and RFR models, with an
of 0.9852, an RMSE of 2.4301%, and a MAE of 2.0250% on the test set. The comparison with the SVR, RFR, and BPNN models, it was revealed that automatically mining a sufficient number of features from the E-nose data using the 1DCNN backbone significantly improved detection performance.
The 10-channel input was submitted to the proposed 1DCNN-RFR framework consisting of the 1DCNN backbone and the RFR. The 1DCNN backbone in the 1DCNN-RFR framework used the model parameters of the trained 1DCNN framework at 920 epochs. The training stage of the RFR in the 1DCNN-RFR framework was the same as that of the RFR model. Through the grid search skill, the max. depth and the min. samples split were set to 5 and 35, respectively. As shown in
Table 5, the proposed 1DCNN-RFR framework achieved a better performance than all other models, with an
of 0.9977, an RMSE of 0.9491%, and a MAE of 0.4619% on the test set. These results indicated that the strong prediction ability of the RFR improved the regression performance in the 1DCNN-RFR framework. The relationships between the predicted adulterated proportions by the five models and the corresponding actual adulterated proportions are shown in
Figure 7. The x-axis represents the sequence number of the tested samples, and the black and red points along the y-axis represent the actual and predicted proportions, respectively.
Figure 7 intuitively illustrates that the predictive performances of models using the 1DCNN backbone to extract features were significantly better than those that did not use the 1DCNN backbone. The proposed 1DCNN-RFR framework achieved the best predictions and predicted almost all adulterated proportions precisely.
4.2.2. Experiment B
In practical applications, the number of samples will probably be much more limited than in Experiment A. Thus, Experiment B used a smaller number of training samples to further evaluate the generalization performance of the models. Dataset B was used in this experiment and was divided into the training set (data from the first 3 days, 63 samples) and the test set (data from the remaining 7 days, 147 samples). For the SVR, RFR, and BPNN models, the training set was expressed as a 630 × 10 matrix and the test set was expressed as a 1470 × 10 matrix. For the 1DCNN and 1DCNN-RFR frameworks, the training set and the test set were expressed as a 63 × 10 × 100 matrix and a 147 × 10 × 100 matrix, respectively.
All the experimental steps were the same as those of Experiment A, except for the 1DCNN where the batch size was adjusted from 49 to 21, while the other parameters were left unchanged. The best parameters of the SVR and RFR models were a penalty factor of 500, gamma of 0.1, max. depth of 3, and min. samples split of 21. The max. depth and min. samples split of the RFR in the 1DCNN-RFR framework were set to 3 and 7, respectively. The training loss of the 1DCNN during training is shown in
Figure 8. The 1DCNN framework converged after 980 epochs. The parameters at 980 epochs were saved and used to evaluate the performance of the 1DCNN framework on the test set. The test set regression results from the five models (the SVR model, RFR model, BPNN model, 1DCNN framework, and 1DCNN-RFR framework) are shown in
Table 6. The prediction performances of the SVR and RFR models in Experiment B were much worse than those of Experiment A. The BPNN model, the 1DCNN framework, and the proposed 1DCNN-RFR framework also suffered a slight reduction in performance. Even so, the 1DCNN framework and the 1DCNN-RFR framework performed much better than the SVR and RFR models. The 1DCNN-RFR model still worked best and obtained a good result with an
of 0.9858, an RMSE of 2.3849%, and a MAE of 1.1625% on the test set. The regression results in Experiment B further demonstrated the superiority of the proposed 1DCNN-RFR framework, which performed well despite the much smaller sample size. The relationships between the predicted adulterated proportions of the five models and the actual adulterated proportions are shown in
Figure 9. These relationships showed that the SVR and RFR models, which did not use the 1DCNN backbone and were unable to extract a sufficient number of features, had extremely poor prediction results. The proposed 1DCNN-RFR framework performed best.