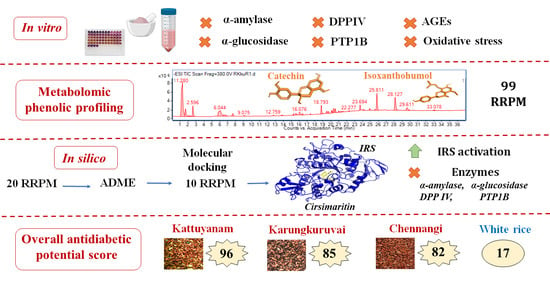
Multi-Mechanistic In Vitro Evaluation of Antihyperglycemic, Antioxidant and Antiglycation Activities of Three Phenolic-Rich Indian Red Rice Genotypes and In Silico Evaluation of Their Phenolic Metabolites
Abstract
:1. Introduction
2. Methodology
2.1. Experimental Samples
2.2. Chemicals and Reagents
2.3. Sample Storage and Phenolic Extraction
2.4. In Vitro Antihyperglycemic Enzyme Assays
2.4.1. Alpha-Amylase Inhibitory Assay
2.4.2. Alpha-Glucosidase Inhibitory Assay
2.4.3. Dipeptidyl Peptidase IV Inhibitory Assay
2.4.4. Protein Tyrosine Phosphatase 1 B Inhibitory Assay
2.5. Antioxidant Assays
2.5.1. DPPH Radical Scavenging Assay
2.5.2. FRAP Assay
2.5.3. ABTS Assay
2.6. Advanced Glycated End Products Inhibitory Potential
2.7. Total Phenolics and Total Flavonoids Analysis
2.8. Q-TOF-LC-MS/MS Analysis for Phenolics Identification
2.9. In Silico Analysis of Red Rice Phenolic Metabolites (RRPM)
2.9.1. ADME Analysis
2.9.2. Molecular Docking Analysis
2.10. Assessment of Overall Antidiabetic Potential Using AHCI and OGRCI
2.11. Statistical Methods
3. Results and Discussion
3.1. In Vitro Antihyperglycemic Activities of Red Rice Extracts
3.1.1. Alpha-Amylase and Glucosidase Inhibitory Activities
3.1.2. Dipeptidyl Peptidase IV Inhibitory Activity
3.1.3. Protein Tyrosine Phosphatase 1 B Inhibitory Activity
3.2. In Vitro Antioxidant Activities of Red Rice Extracts
3.3. Advanced Glycation end Products Inhibitory Activity of Red Rice Extracts
3.4. Total Phenolic and Total Flavonoid Contents of Red Rice Extracts
3.5. Metabolomics Phenolic Profiling
3.5.1. The Unique Phenolic Fingerprint of Red Rice Varieties
3.5.2. Relative Quantification of Red Rice Phenolic Metabolites
3.5.3. Principal Component Analysis of the Red Rice Genotypes with Phenolic Metabolites
3.5.4. Reported In Vitro and In Vivo Antihyperglycemic Activity of the Identified Abundant Red Rice Phenolic Metabolites
3.5.5. Reported Antioxidant and Antiglycation Activity of Identified Abundant Red Rice Phenolic Metabolites
3.6. In Silico Analysis of Red Rice Phenolic Metabolites
3.6.1. ADME Screening of Red Rice Phenolic Metabolites
3.6.2. Molecular Docking of Red Rice Phenolic Metabolites with Glucose Regulatory Target Proteins
3.7. Overview of the Antidiabetic Potential of the Red Rice Cultivars
- Delaying gastric emptying leading to the reduction of post-prandial blood glucose by inhibiting carbohydrate metabolizing enzymes implicated in diabetes;
- Increasing insulin secretion by preventing the degradation of incretin hormones through DPP IV inhibition;
- Improving insulin sensitivity and insulin signaling by inhibiting PTP1B enzyme;
- Preventing the development and progression of diabetes and its vascular complications by reducing oxidative stress;
- Preventing the development and progression of diabetes and its vascular complications by inhibiting the formation of AGEs;
- Increasing uptake of glucose by activating the insulin receptor complex.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Williams, R.; Karuranga, S.; Malanda, B.; Saeedi, P.; Basit, A.; Besançon, S.; Bommer, C.; Esteghamati, A.; Ogurtsova, K.; Zhang, P.; et al. Global and regional estimates and projections of diabetes-related health expenditure: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2020, 162, 108072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerf, M.E. Beta Cell Dysfunction and Insulin Resistance. Front. Endocrinol. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhury, A.; Duvoor, C.; Reddy Dendi, V.S.; Kraleti, S.; Chada, A.; Ravilla, R.; Marco, A.; Shekhawat, N.S.; Montales, M.T.; Kuriakose, K.; et al. Clinical Review of Antidiabetic Drugs: Implications for Type 2 Diabetes Mellitus Management. Front. Endocrinol. 2017, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Hussain, A.; Misra, A. Diabetes and COVID-19: Evidence, current status and unanswered research questions. Eur. J. Clin. Nutr. 2020, 74, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Alkhatib, A.; Tsang, C.; Tiss, A.; Bahorun, T.; Arefanian, H.; Barake, R.; Tuomilehto, J. Functional foods and lifestyle approaches for diabetes prevention and management. Nutrients 2017, 9, 1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herforth, A.; Arimond, M.; Álvarez-Sánchez, C.; Coates, J.; Christianson, K.; Muehlhoff, E. A Global Review of Food-Based Dietary Guidelines. Adv. Nutr. 2019, 10, 590–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, A.; Pan, A.; Malik, V.; Sun, Q. White rice consumption and risk of type 2 diabetes: Meta-analysis and systematic review. BMJ 2019, 2001, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozuka, C.; Yabiku, K.; Takayama, C.; Matsushita, M.; Shimabukuro, M.; Masuzaki, H. Natural food science based novel approach toward prevention and treatment of obesity and type 2 diabetes: Recent studies on brown rice and γ-oryzanol. Obes. Res. Clin. Pract. 2013, 7, e165–e172. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.-Y.; Xu, W.-J.; Tao, J.; Zhang, J.-H.; Zhou, X.-R.; Yuan, G.; Yang, Y.; Zhang, J.; Zhang, H.-Y.; Xu, Q.; et al. Glycemic index, glycemic load, and glycemic response to pomelo in patients with type 2 diabetes. Curr. Med. Sci. 2017, 37, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Lankatillake, C.; Huynh, T.; Dias, D.A. Understanding glycemic control and current approaches for screening antidiabetic natural products from evidence-based medicinal plants. Plant Methods 2019, 15, 1–35. [Google Scholar] [CrossRef] [Green Version]
- Haldipur, A.C.; Srividya, N. A comparative evaluation of in vitro antihyperglycemic potential of Bamboo seed rice (Bambusa arundinacea) and Garudan samba (Oryza sativa): An integrated metabolomics, enzymatic and molecular docking approach. J. Cereal Sci. 2021, 99, 103200. [Google Scholar] [CrossRef]
- Armengol, G.D.; Hayfron-Benjamin, C.F.; Born, B.-J.H.V.D.; Galenkamp, H.; Agyemang, C. Microvascular and macrovascular complications in type 2 diabetes in a multi-ethnic population based in Amsterdam. The HELIUS study. Prim. Care Diabetes 2021, 15, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Spasov, A.A.; Solov’Eva, O.A.; Kuznetsova, V.A. Protein Glycation During Diabetes Mellitus and the Possibility of its Pharmacological Correction. Pharm. Chem. J. 2017, 51, 429–433. [Google Scholar] [CrossRef]
- Asgar, M.D.A. Anti-diabetic potential of phenolic compounds: A review. Int. J. Food Prop. 2013, 16, 91–103. [Google Scholar] [CrossRef]
- Khan, M.; Liu, H.; Wang, J.; Sun, B. Inhibitory effect of phenolic compounds and plant extracts on the formation of advance glycation end products: A comprehensive review. Food Res. Int. 2020, 130, 108933. [Google Scholar] [CrossRef]
- Deng, G.-F.; Xu, X.-R.; Zhang, Y.; Li, D.; Gan, R.-Y.; Li, H.-B. Phenolic Compounds and Bioactivities of Pigmented Rice. Crit. Rev. Food Sci. Nutr. 2013, 53, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.; Dutta, H.; Saikia, D.; Mahanta, C. Quality characterisation and estimation of phytochemicals content and antioxidant capacity of aromatic pigmented and non-pigmented rice varieties. Food Res. Int. 2012, 46, 334–340. [Google Scholar] [CrossRef]
- Kaur, P.; Singh, N.; Pal, P.; Kaur, A. Variation in composition, protein and pasting characteristics of different pigmented and non-pigmented rice (Oryza sativa L.) grown in Indian Himalayan region. J. Food Sci. Technol. 2018, 55, 3809–3820. [Google Scholar] [CrossRef]
- Meera, K.; Smita, M.; Haripriya, S.; Sen, S. Varietal influence on antioxidant properties and glycemic index of pigmented and non-pigmented rice. J. Cereal Sci. 2019, 87, 202–208. [Google Scholar] [CrossRef]
- Choudhury, P.; Dutta, K.N.; Singh, A.; Malakar, D.; Pillai, M.; Talukdar, N.C.; Samanta, S.K.; Devi, R. Assessment of nutritional value and quantitative analysis of bioactive phytochemicals through targeted LC-MS/MS method in selected scented and pigmented rice varietals. J. Food Sci. 2020, 85, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Haldipur, A.C.; Srividya, N. In vitro glycemic response of indigenous pigmented rice cultivars from south India and in-fluence of different carbohydrate components. Curr. Res. Nutr. Food Sci. J. 2020, 8, 815–828. [Google Scholar] [CrossRef]
- Bordiga, M.; Gomez-Alonso, S.; Locatelli, M.; Travaglia, F.; Coïsson, J.D.; Hermosin-Gutierrez, I.; Arlorio, M. Phenolics char-acterization and antioxidant activity of six different pigmented Oryza sativa L. cultivars grown in Piedmont (Italy). Food Res. Int. 2014, 65, 282–290. [Google Scholar] [CrossRef]
- Visvanathan, R.; Jayathilake, C.; Liyanage, R. A simple microplate-based method for the determination of α-amylase activity using the glucose assay kit (GOD method). Food Chem. 2016, 211, 853–859. [Google Scholar] [CrossRef]
- Kang, W.Y.; Song, Y.L.; Zhang, L. α-Glucosidase inhibitory and antioxidant properties and antidiabetic activity of Hypericum ascyron L. Med. Chem. Res. 2011, 20, 809–816. [Google Scholar] [CrossRef]
- Fan, J.; Johnson, M.H.; Lila, M.A.; Yousef, G.; De Mejia, E.G. Berry and Citrus Phenolic Compounds Inhibit Dipeptidyl Peptidase IV: Implications in Diabetes Management. Evid.-Based Complement. Altern. Med. 2013, 2013, 479505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, L.; Cao, J.; Duan, L.; Tang, Y.; Zhao, Y. Protein tyrosine phosphatase 1B (PTP1B) and α-glucosidase inhibitory activities of Schisandra chinensis (Turcz.) Baill. J. Funct. Foods 2014, 9, 264–270. [Google Scholar] [CrossRef]
- Yu, L.; Haley, S.; Perret, J.; Harris, M.; Wilson, J.; Qian, M. Free Radical Scavenging Properties of Wheat Extracts. J. Agric. Food Chem. 2002, 50, 1619–1624. [Google Scholar] [CrossRef]
- Jimenez-Alvarez, D.; Giuffrida, F.; Vanrobaeys, F.; Golay, P.A.; Cotting, C.; Lardeau, A.; Keely, B.J. High-Throughput Methods To Assess Lipophilic and Hydrophilic Antioxidant Capacity of Food Extracts in Vitro. J. Agric. Food Chem. 2008, 56, 3470–3477. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Sharma, S.D.; Pandey, B.N.; Mishra, K.P.; Sivakami, S. Amadori product and age formation during nonenzymatic glycosylation of bovine serum albumin in vitro. J. Biochem. Mol. Biol. Biophys. 2002, 6, 233–242. [Google Scholar]
- Awasthi, S.; Saraswathi, N.T. Silybin, a flavonolignan from milk thistle seeds, restrains the early and advanced glycation end product modification of albumin. RSC Adv. 2015, 5, 87660–87666. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, J.; Shen, J.; Silva, A.; Dennis, D.A.; Barrow, C.J. A Simple 96-Well Microplate Method for Estimation of Total Polyphenol Content in Seaweeds. J. Appl. Phycol. 2006, 18, 445–450. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-O.; Chun, O.K.; Kim, Y.J.; Moon, A.H.-Y.; Lee, C.Y. Quantification of Polyphenolics and Their Antioxidant Capacity in Fresh Plums. J. Agric. Food Chem. 2003, 51, 6509–6515. [Google Scholar] [CrossRef]
- Brayer, G.D.; Sidhu, G.; Maurus, R.; Rydberg, E.H.; Braun, C.; Wang, Y.; Withers, S.G. Subsite mapping of the human pan-creatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry 2000, 39, 4778–4791. [Google Scholar] [CrossRef]
- Shen, X.; Saburi, W.; Gai, Z.; Kato, K.; Ojima-Kato, T.; Yu, J.; Komoda, K.; Kido, Y.; Matsui, H.; Mori, H.; et al. Structural analysis of the α-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1382–1391. [Google Scholar] [CrossRef]
- Ha, M.T.; Park, D.H.; Shrestha, S.; Kim, M.; Kim, J.A.; Woo, M.H.; Choi, J.S.; Min, B.S. PTP1B inhibitory activity and molecular docking analysis of stilbene derivatives from the rhizomes of Rheum undulatum L. Fitoterapia 2018, 131, 119–126. [Google Scholar] [CrossRef]
- Pradhan, J.; Panchawat, S. Molecular Docking Studies and Pharmacophore Modeling of Some Insulin Mimetic Agents from Herbal Sources: A Rational Approach towards Designing of Orally Active Insulin Mimetic Agents. Curr. Tradit. Med. 2020, 6, 121–133. [Google Scholar] [CrossRef]
- Balakrishnan, J.; Thamilarasan, S.K.; Ravi, M.S.; Pugazhendhi, A.; Perumal, V.; Padikasan, I.A. Comparison of phytochemicals, antioxidant and hypoglycemic activity of four different Brown rice varieties. Biocatal. Agric. Biotechnol. 2019, 21, 101351. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Menichini, F. Natural Products as α-Amylase and α-Glucosidase Inhibitors and their Hypoglycaemic Potential in the Treatment of Diabetes: An Update. Mini Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef]
- Hatanaka, T.; Inoue, Y.; Arima, J.; Kumagai, Y.; Usuki, H.; Kawakami, K.; Kimura, M.; Mukaihara, T. Production of dipeptidyl peptidase IV inhibitory peptides from defatted rice bran. Food Chem. 2012, 134, 797–802. [Google Scholar] [CrossRef]
- Hatanaka, T.; Uraji, M.; Fujita, A.; Kawakami, K. Anti-oxidation activities of rice-derived peptides and their inhibitory effects on dipeptidyl peptidase-IV. Int. J. Pept. Res. Ther. 2015, 21, 479–485. [Google Scholar] [CrossRef]
- Liu, R.; Cheng, J.; Wu, H. Discovery of Food-Derived Dipeptidyl Peptidase IV Inhibitory Peptides: A Review. Int. J. Mol. Sci. 2019, 20, 463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barik, S.; Russell, W.; Dehury, B.; Cruickshank, M.; Moar, K.; Thapa, D.; Hoggard, N. Dietary phenolics other than anthocyanins inhibit PTP1B; an in vitro and in silico validation. Proc. Nutr. Soc. 2019, 78. [Google Scholar] [CrossRef] [Green Version]
- Eleftheriou, P.; Geronikaki, A.; Petrou, A. PTP1b Inhibition, A Promising Approach for the Treatment of Diabetes Type II. Curr. Top. Med. Chem. 2019, 19, 246–263. [Google Scholar] [CrossRef]
- Munteanu, I.G.; Apetrei, C. Analytical Methods Used in Determining Antioxidant Activity: A Review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef]
- Fidrianny, I.; Puspitaningrum, D.A.; Ruslan, K. Antioxidant capacities of various grains extracts of three kinds of rice grown in Central Java-Indonesia. Int. J. Pharm. Phytochem. Res. 2016, 8, 997–1002. [Google Scholar]
- Dos Santos, J.M.; Tewari, S.; Mendes, R.H. The Role of Oxidative Stress in the Development of Diabetes Mellitus and Its Complications. J. Diabetes Res. 2019, 2019, 4189813. [Google Scholar] [CrossRef] [Green Version]
- Daiponmak, W.; Senakun, C.; Siriamornpun, S. Antiglycation capacity and antioxidant activities of different pigmented Thai rice. Int. J. Food Sci. Technol. 2014, 49, 1805–1810. [Google Scholar] [CrossRef]
- Sumczynski, D.; Kotásková, E.; Družbíková, H.; Mlcek, J. Determination of contents and antioxidant activity of free and bound phenolics compounds and in vitro digestibility of commercial black and red rice (Oryza sativa L.) varieties. Food Chem. 2016, 211, 339–346. [Google Scholar] [CrossRef]
- Quero, J.; Mármol, I.; Cerrada, E.; Rodríguez-Yoldi, M.J. Insight into the potential application of polyphenol-rich dietary intervention in degenerative disease management. Food Funct. 2020, 11, 2805–2825. [Google Scholar] [CrossRef]
- Hossain, M.K.; Dayem, A.A.; Han, J.; Yin, Y.; Kim, K.; Saha, S.K.; Yang, G.-M.; Choi, H.Y.; Cho, S.-G. Molecular Mechanisms of the Anti-Obesity and Anti-Diabetic Properties of Flavonoids. Int. J. Mol. Sci. 2016, 17, 569. [Google Scholar] [CrossRef] [Green Version]
- Vinayagam, R.; Jayachandran, M.; Xu, B. Antidiabetic Effects of Simple Phenolic Acids: A Comprehensive Review. Phytother. Res. 2016, 30, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Robards, K.; Helliwell, S.; Blanchard, C. The distribution of phenolic acids in rice. Food Chem. 2004, 87, 401–406. [Google Scholar] [CrossRef]
- Liu, L.; Guo, J.; Zhang, R.; Wei, Z.; Deng, Y.; Guo, J.; Zhang, M. Effect of degree of milling on phenolic profiles and cellular antioxidant activity of whole brown rice. Food Chem. 2015, 185, 318–325. [Google Scholar] [CrossRef]
- Gong, E.S.; Luo, S.J.; Li, T.; Liu, C.M.; Zhang, G.W.; Chen, J.; Zeng, Z.C.; Liu, R.H. Phytochemical profiles and antioxidant activity of brown rice varieties. Food Chem. 2017, 227, 432–443. [Google Scholar] [CrossRef]
- Ziegler, V.; Ferreira, C.D.; Hoffmann, J.F.; Chaves, F.C.; Vanier, N.L.; De Oliveira, M.; Elias, M.C. Cooking quality properties and free and bound phenolics content of brown, black, and red rice grains stored at different temperatures for six months. Food Chem. 2018, 242, 427–434. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Fu, T.; Sheng, Y.; Zhang, S.; Zhang, Y.; Jiang, Y.; Yu, M.; Zhang, D. Effect of storage on metabolites of brown rice. J. Sci. Food Agric. 2020, 100, 4364–4377. [Google Scholar] [CrossRef]
- Yamashita, M.; Fukizawa, S.; Nonaka, Y. Hop-derived prenylflavonoid isoxanthohumol suppresses insulin resistance by changing the intestinal microbiota and suppressing chronic inflammation in high fat diet-fed mice. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1537–1547. [Google Scholar]
- Hanchang, W.; Khamchan, A.; Wongmanee, N.; Seedadee, C. Hesperidin ameliorates pancreatic β-cell dysfunction and apoptosis in streptozotocin-induced diabetic rat model. Life Sci. 2019, 235, 116858. [Google Scholar] [CrossRef]
- Jayachandran, M.; Wu, Z.; Ganesan, K.; Khalid, S.; Chung, S.; Xu, B. Isoquercetin upregulates antioxidant genes, suppresses inflammatory cytokines and regulates AMPK pathway in streptozotocin-induced diabetic rats. Chem. Biol. Interact. 2019, 303, 62–69. [Google Scholar] [CrossRef]
- Samarghandian, S.; Azimi-Nezhad, M.; Farkhondeh, T. Catechin Treatment Ameliorates Diabetes and Its Complications in Streptozotocin-Induced Diabetic Rats. Dose-Response 2017, 15, 1559325817691158. [Google Scholar] [CrossRef]
- Nazir, N.; Zahoor, M.; Ullah, R.; Ezzeldin, E.; Mostafa, G.A.E. Curative Effect of Catechin Isolated from Elaeagnus umbellata Thunb. Berries for Diabetes and Related Complications in Streptozotocin-Induced Diabetic Rats Model. Molecules 2021, 26, 137. [Google Scholar] [CrossRef] [PubMed]
- Dhanya, R.; Jayamurthy, P. In vitro evaluation of antidiabetic potential of hesperidin and its aglycone hesperetin under oxidative stress in skeletal muscle cell line. Cell Biochem. Funct. 2020, 38, 419–427. [Google Scholar] [CrossRef]
- Potaniec, B.; Grabarczyk, M.; Stompor, M.; Szumny, A.; Zieliński, P.; Żołnierczyk, A.K.; Anioł, M. Antioxidant activity and spectroscopic data of isoxanthohumol oxime and related compounds. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 118, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Bao, Y.; Zhu, Y.; Liu, S.; Peng, Z.; Zhang, Y.; Zhou, G. Isorhamnetin, Hispidulin, and Cirsimaritin Identified in Tamarix ramosissima Barks from Southern Xinjiang and Their Antioxidant and Antimicrobial Activities. Molecules 2019, 24, 390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Chen, Z.; Yan, M.; He, P.; Chen, Z.; Dai, H. The protective role of isorhamnetin on human brain microvascular en-dothelial cells from cytotoxicity induced by methylglyoxal and oxygen-glucose deprivation. J. Neurochem. 2016, 136, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Orsini, F.; Vovk, I.; Glavnik, V.; Jug, U.; Corradini, D. HPTLC, HPTLC-MS/MS and HPTLC-DPPH methods for analyses of flavonoids and their antioxidant activity in Cyclanthera pedata leaves, fruits and dietary supplement. J. Liq. Chromatogr. Relat. Technol. 2019, 42, 290–301. [Google Scholar] [CrossRef]
- Zhou, Q.; Cheng, K.-W.; Gong, J.; Li, E.T.; Wang, M. Apigenin and its methylglyoxal-adduct inhibit advanced glycation end products-induced oxidative stress and inflammation in endothelial cells. Biochem. Pharmacol. 2019, 166, 231–241. [Google Scholar] [CrossRef]
- Vongsak, B.; Mangmool, S.; Gritsanapan, W. Antioxidant Activity and Induction of mRNA Expressions of Antioxidant Enzymes in HEK-293 Cells of Moringa oleifera Leaf Extract. Planta Medica 2015, 81, 1084–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Bassey, A.P.; Khan, I.A.; Huang, M.; Zhang, X. Inhibitory mechanism of catechins against advanced glycation end products of glycated myofibrillar protein through anti-aggregation and anti-oxidation. LWT 2021, 147, 111550. [Google Scholar] [CrossRef]
- Liu, D.; Cao, X.; Kong, Y.; Mu, T.; Liu, J. Inhibitory mechanism of sinensetin on α-glucosidase and non-enzymatic glycation: Insights from spectroscopy and molecular docking analyses. Int. J. Biol. Macromol. 2021, 166, 259–267. [Google Scholar] [CrossRef]
- Wilcox, G. Insulin, and insulin resistance. Clin. Biochem. Rev. 2005, 26, 19. [Google Scholar] [PubMed]
- Manukumar, H.M.; Kumar, J.S.; Chandrasekhar, B.; Raghava, S.; Umesha, S. Evidences for diabetes and insulin mimetic activity of medicinal plants: Present status and future prospects. Crit. Rev. Food Sci. Nutr. 2017, 57, 2712–2729. [Google Scholar] [CrossRef]
- Hua, F.; Zhou, P.; Wu, H.-Y.; Chu, G.-X.; Xie, Z.-W.; Bao, G.-H. Inhibition of α-glucosidase and α-amylase by flavonoid glycosides from Lu’an GuaPian tea: Molecular docking and interaction mechanism. Food Funct. 2018, 9, 4173–4183. [Google Scholar] [CrossRef]
- Cai, Y.; Wu, L.; Lin, X.; Hu, X.; Wang, L. Phenolic profiles and screening of potential α-glucosidase inhibitors from Polygonum aviculare L. leaves using ultra-filtration combined with HPLC-ESI-qTOF-MS/MS and molecular docking analysis. Ind. Crop. Prod. 2020, 154, 112673. [Google Scholar] [CrossRef]
- Sachan, R.; Kumar, V.; Sharma, K.; Abbasi, F.; Anwar, F. Antihyperglycemic activity with DPP-IV inhibition of Prunus amygdalus extract: Investigation via experimental validation and molecular docking. Authorea Prepr. 2020. [Google Scholar] [CrossRef]
- Sarikurkcu, C.; Sahinler, S.S.; Husunet, M.T.; Istifli, E.S.; Tepe, B. Two endemic Onosma species (O. sieheana and O. stenoloba): A comparative study including docking data on biological activity and phenolic composition. Ind. Crop. Prod. 2020, 154, 112656. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, B.; Tan, C.; Huang, Q. α-Glucosidase inhibitors: Consistency of in silico docking data with in vitro inhibitory data and inhibitory effect prediction of quercetin derivatives. Food Funct. 2019, 10, 6312–6321. [Google Scholar] [CrossRef] [PubMed]
- Jhong, C.H.; Riyaphan, J.; Lin, S.H.; Chia, Y.C.; Weng, C.F. Screening alpha-glucosidase and alpha-amylase inhibitors from natural compounds by molecular docking in silico. Biofactors 2015, 41, 242–251. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Chaturvedi, N.; Singh, A.; Mishra, A. Characterization, inhibitory activity and mechanism of polyphenols from faba bean (gallic-acid and catechin) on α-glucosidase: Insights from molecular docking and simulation study. Prep. Biochem. Biotechnol. 2020, 50, 123–132. [Google Scholar] [CrossRef]
- Kempegowda, P.K.; Zameer, F.; Murari, S.K. Delineating antidiabetic proficiency of catechin from Withania somnifera and its Inhibitory action on dipeptidyl peptidase-4 (DPP-4). Biomed. Res. 2018, 29, 3192–3200. [Google Scholar] [CrossRef] [Green Version]
- Kuban-Jankowska, A.; Kostrzewa, T.; Musial, C.; Barone, G.; Bosco, G.L.; Celso, F.L.; Gorska-Ponikowska, M. Green Tea Catechins Induce Inhibition of PTP1B Phosphatase in Breast Cancer Cells with Potent Anti-Cancer Properties: In Vitro Assay, Molecular Docking, and Dynamics Studies. Antioxidants 2020, 9, 1208. [Google Scholar] [CrossRef] [PubMed]
- Sherafati, M.; Mohammadi-Khanaposhtani, M.; Moradi, S.; Asgari, M.S.; Najafabadipour, N.; Faramarzi, M.A.; Mahdavi, M.; Biglar, M.; Larijani, B.; Hamedifar, H.; et al. Design, synthesis and biological evaluation of novel phthalimide-Schiff base-coumarin hybrids as potent α-glucosidase inhibitors. Chem. Pap. 2020, 74, 4379–4388. [Google Scholar] [CrossRef]
- Hong, B.; He, J.; Fan, C.; Tang, C.; Le, Q.; Bai, K.; Niu, S.; Xiao, M. Synthesis and Biological Evaluation of Analogues of Butyrolactone I as PTP1B Inhibitors. Mar. Drugs 2020, 18, 526. [Google Scholar] [CrossRef] [PubMed]






| Red Rice Genotypes | α-Amylase IC50 (µg/mL) | α-Glucosidase IC50 (µg/mL) | DPP IV IC50 (µg/mL) | PTP 1B IC50 (µg/mL) |
|---|---|---|---|---|
| Kattuyanam | 2.75 ± 0.09 a | 22.39 ± 0.08 c | 2.18 ± 0.08 c | 31.39 ± 0.18 c |
| Chennangi | 5.71 ± 0.07 c | 18.51 ± 0.13 b | 5.68 ± 0.14 d | 38.57 ± 0.21 d |
| Karungkuruvai | 4.28 ± 0.11 b | 24.62 ± 0.12 d | 1.66 ± 0.06 b | 30.89 ± 0.16 b |
| Sona masuri (White rice) | 58.64 ± 1.15 d | 82.35 ± 0.19 e | 72.57 ± 1.30 e | ND |
| Positive controls | 2.67 ± 0.08 a (Acarbose) | 11.24 ± 0.12 a (Acarbose) | 1.19 ± 0.14 a (Sitagliptin) | 28.41± 0.19 a (Sodium orthovanadate) |
| Red Rice Genotypes | DPPH RSA (% Inhibition) | FRAP (µmol Fe2+/g) | TEACABTS (µmol TE/g) | AGEs Formation Inhibition (% Inhibition) |
|---|---|---|---|---|
| Kattuyanam | 94.89 ± 0.16 e | 118.68 ± 0.30 d | 24.48 ± 0.21 e | 68.17 ± 0.43 c |
| Chennangi | 92.11 ± 0.08 d | 99.68 ± 0.18 b | 7.56 ± 0.03 b | 55.30 ± 0.21 b |
| Karungkuruvai | 86.31 ± 0.21 b | 112.65 ± 0.20 c | 9.94 ± 0.04 c | 65.90 ± 0.79 c |
| Sona masuri (White rice) | 5.21 ± 0.08 a | 7.61 ± 0.21 a | 0.67 ± 0.03 a | 5.35 ± 0.19 a |
| Positive controls | 89.16 ± 0.11 c (Ascorbic acid) | 123.27 ± 0.16 e (Ascorbic acid) | 18.71 ± 0.06 d (Butylated hydroxytoluene) | 91.21 ± 0.81 d (Aminoguanidine) |
| Red Rice Genotypes | Total Phenolics (mg GAE/100 g) | Total Flavonoids (mg QE/100 g) |
|---|---|---|
| Kattuyanam | 731.41 ± 0.25 d | 246.13 ± 0.19 d |
| Chennangi | 499.01 ± 0.22 b | 243.14 ± 0.55 c |
| Karungkuruvai | 510.96 ± 0.41 c | 235.24 ± 0.11 b |
| Sona masuri (white rice) | 92.34 ± 0.56 a | 73.88 ± 0.75 a |
| Red Rice Phenolic Metabolites | Binding Energy (kcal/mol) with Target Proteins | ||||
|---|---|---|---|---|---|
| α-Amylase | α-Glucosidase | DPP IV | PTP 1B | IRS | |
| 3′,4′,7-Trihydroxyisoflavone | −5.27 | −6.84 | −6.89 | −7.89 | −5.05 |
| 3-methylcatechol | −4.44 | −5.43 | −5.17 | −5.14 | −4.33 |
| 3′-O-Methylviolanone | −7.13 | −7.84 | −6.95 | −7.31 | −7.00 |
| 4-Hydroxybenzaldehyde | −4.14 | −4.76 | −4.30 | −4.99 | −4.32 |
| Benzoic acid | −4.66 | −4.54 | −5.00 | −4.87 | −3.25 |
| Catechin | −6.72 | −8.67 | −7.81 | −7.17 | −7.41 |
| Cirsimaritin | −6.41 | −7.86 | −9.79 | −7.46 | −8.00 |
| Dihydroquercetin | −7.91 | −7.91 | −7.43 | −6.89 | −7.25 |
| Isorhamnetin | −5.62 | −7.89 | −7.75 | −7.65 | −6.93 |
| Isoxanthohumol | −7.64 | −9.26 | −9.49 | −8.72 | −8.42 |
| Red Rice Phenolic Metabolites | α-Amylase | α-Glucosidase | DPPIV | PTP1B | IRS |
|---|---|---|---|---|---|
| 3′,4′,7-Trihydroxyisoflavone | GLU233, ASP197 (H bonds), ASP300 (Pi anion) | ARG400 (H bond), ASP333 (Halogen, Pi-anion, Pi sulfur), PHE147 (Pi sulfur), TYR65 (alkyl), PHE166 (Pi sigma), GLU271 (H bond) | TYR666 (H bond), TYR662 TYR631, PHE357 (Alkyl), SER209, GLU205, GLU206, TYR547 (H bond) | CYS215, ASP181, SER216 (C-H bonds), TYR46 (C-H bond & Pi-alkyl bond), ARG221 (C-H bond & alkyl interaction), GLN262 (halogen (fluorine bond), ILE219, PHE182 (Pi alkyl), ALA217 (Pi-Alkyl and Pi-Sigma) | ASP 1083 (l H bond), MET1079 (H bond), GLU1077 (Halogen fluorine) |
| 3′-O-Methylviolanone | GLU233 (H bond) | GLY228, PHE147, PHE166, ARG400, ASP333, PHE297 (Van der Waals) | TYR631, TRP659, VAL656, VAL711, ASN710, GLU205 (Van der Waal) ARG358, SER630, TYR662 (H bond) TYR547, SER209 (C-H bond) GLU206 (Pi-donor H bond), PHE357 (Pi-alkyl bond) | PHE182, ILE219, GLN262, ARG221 (Van der Waals), ALA217 (Pi donor H bond), CYS215 (Pi alkyl bond), ASP181 (Pi anion) | GLU1077 (Conv. H bond), ASP1083 (Carbon H bond), MET1079 (Pi-alkyl bond) |
| Catechin | GLU233, ASP197 (H bonds), ASP300 (Pi anion) | GLY228 (H bond), ARG400 (Pi cation), GLU271, PHE147, ASP202, HIS105 (Van der Waals) | GLU205, GLU206, SER209, TYR662, SER630 (H bonds), TYR547 (Pi donor H bond) | PHE182 (Pi alkyl), SER216, CYS215, TYR46 (C-H bond), ILE219 (Alkyl and C-H bond) | ASP1083, MET1079, GLU1077, ASP1150 (Conventional H bonds) |
| Cirsimaritin | GLU233 (H bond), ASP300 (Pi anion) | HIS105, ASP202, GLY228, ARG400 (H bonds), PHE166, TYR65 (Pi-Pi stacked), PHE297 (Alkyl) | GLU205, SER209, ARG358, TYR547, SER630 (H bonds), ASN710, HIS740 (C-H bond), TYR631, VAL656, TYR662, VAL711 (Alkyl bonds), GLU206 (Pi anion) | ARG221, CYS215 (H bond), (Van der Waals), ASP181 (Pi anion bond), TYR46 (Pi-Pi stacked), ALA217 (Pi alkyl bond) | MET1079 (Conv. H bond), ASP1083 (Carbon H bond) |
| Dihydroquercetin | GLU233 (H bond), ASP300 (Pi anion) | GLY228 (H bond), PHE147, THR203, GLU271, ASP62 (Van der Waals), ASP202, ARG200, (H bond), ASP333 (Pi anion), TYR65 (Pi-Pi T shaped) | GLU206 (Pi anion), ARG358, TYR631, ASN710 (Van der Waals), GLU205, SER209, SER630, TYR662, TYR666 (H bond), TYR547 (Pi donor H bond), PHE357 (Pi-Pi T shaped) | Nil | GLU1077, MET1079 (Conv. H bond) |
| Isorhamnetin | GLU233, ASP197 (H bonds) | ASP202, GLY228, ASP333, ARG400 (H bond), TYR65, PHE147, (Pi-Pi T shaped), PHE166 (Pi sigma), HIS105 (Pi alkyl) | GLU205, SER209, GLU206 (H bonds), SER630, TYR547 (Pi-donor bonds), TYR631 (Pi alkyl bond) | SER216 (Van der Waals), ARG221 (H bond), PHE182 (Pi sigma), TYR46 (Pi stacked), CYS215 (Pi-alkyl) ASP181 (Pi-anion) | GLU1077, MET1079 (Conv. H bond), LYS1030 (alkyl) |
| Isoxanthohumol | ASP300 (H bond), GLU233 (Pi anion) | GLU271, ASP333 (Pi anion), GLY228 (Pi sigma), PHE147, ASP202 (C-H bond), PHE166 (Alkyl), PHE297 (Pi-Pi T shaped) | SER209, TYR662, TYR666 (H bonds), VAL656 (Pi alkyl), GLU206 (Pi anion), TYR547 (Pi donor), PHE357 (Pi-Pi T shaped bond) | ASP181 (H bond), SER216 (C-H bond), ALA217, CYS215, ARG221 (alkyl), TYR46, PHE182 (Pi-Pi T shaped) | MET1079 (Conventional H bond), ASP1150 (Pi-sigma), ASP1083 (Carbon H bond) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haldipur, A.C.; Srividya, N. Multi-Mechanistic In Vitro Evaluation of Antihyperglycemic, Antioxidant and Antiglycation Activities of Three Phenolic-Rich Indian Red Rice Genotypes and In Silico Evaluation of Their Phenolic Metabolites. Foods 2021, 10, 2818. https://doi.org/10.3390/foods10112818
Haldipur AC, Srividya N. Multi-Mechanistic In Vitro Evaluation of Antihyperglycemic, Antioxidant and Antiglycation Activities of Three Phenolic-Rich Indian Red Rice Genotypes and In Silico Evaluation of Their Phenolic Metabolites. Foods. 2021; 10(11):2818. https://doi.org/10.3390/foods10112818
Chicago/Turabian StyleHaldipur, Ashrita C., and Nagarajan Srividya. 2021. "Multi-Mechanistic In Vitro Evaluation of Antihyperglycemic, Antioxidant and Antiglycation Activities of Three Phenolic-Rich Indian Red Rice Genotypes and In Silico Evaluation of Their Phenolic Metabolites" Foods 10, no. 11: 2818. https://doi.org/10.3390/foods10112818