Restricted Feeding Resets the Peripheral Clocks of the Digestive System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
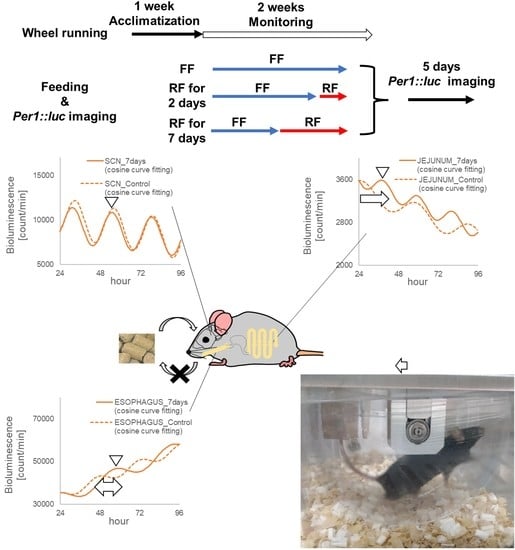
2.1.1. Monitoring the Wheel Running Activity Rhythms of Per1::luc Tg Mice
2.1.2. RF (Restricted Feeding) Conditions
2.1.3. Monitoring the Circadian Rhythms of Each Tissue of Per1::luc Tg Mice
2.1.4. Statistical Analyses
3. Results
3.1. Wheel Running Activity Rhythms during RF
3.2. Monitoring Circadian Rhythms in Each Tissue Sample of the Per1::luc Tg Mice
3.3. Phase Shift of the Bioluminescent Emission Rhythms during RF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tei, H.; Okamura, H.; Shigeyoshi, Y.; Fukuhara, C.; Ozawa, R.; Hirose, M.; Sakaki, Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 1997, 389, 512–516. [Google Scholar] [CrossRef]
- Sun, Z.S.; Albrecht, U.; Zhuchenko, O.; Bailey, J.; Eichele, G.; Lee, C.C. RIGUI, a Putative Mammalian Ortholog of the Drosophila period Gene. Cell 1997, 90, 1003–1011. [Google Scholar] [CrossRef]
- Shearman, L.P.; Zylka, M.J.; Weaver, D.R.; Kolakowski, L.F.; Reppert, S.M. Two period Homologs: Circadian Expression and Photic Regulation in the Suprachiasmatic Nuclei. Neuron 1997, 19, 1261–1269. [Google Scholar] [CrossRef]
- Siepka, S.M.; Yoo, S.-H.; Park, J.; Lee, C.; Takahashi, J.S. Genetics and Neurobiology of Circadian Clocks in Mammals. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Shigeyoshi, Y.; Taguchi, K.; Yamamoto, S.; Takekida, S.; Yan, L.; Tei, H.; Moriya, T.; Shibata, S.; Loros, J.J.; Dunlap, J.C.; et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 1997, 91, 1043–1053. [Google Scholar] [CrossRef]
- Numano, R.; Yamazaki, S.; Umeda, N.; Samura, T.; Sujino, M.; Takahashi, R.-I.; Ueda, M.; Mori, A.; Yamada, K.; Sakaki, Y.; et al. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc. Natl. Acad. Sci. USA 2006, 103, 3716–3721. [Google Scholar] [CrossRef]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.-I.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting Central and Peripheral Circadian Oscillators in Transgenic Rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef]
- Page, A.J. Gastrointestinal Vagal Afferents and Food Intake: Relevance of Circadian Rhythms. Nutrients 2021, 13, 844. [Google Scholar] [CrossRef]
- Grosjean, E.; Simonneaux, V.; Challet, E. Reciprocal Interactions between Circadian Clocks, Food Intake, and Energy Metabolism. Biology 2023, 12, 539. [Google Scholar] [CrossRef]
- Stokkan, K.-A.; Yamazaki, S.; Tei, H.; Sakaki, Y.; Menaker, M. Entrainment of the Circadian Clock in the Liver by Feeding. Science 2001, 291, 490–493. [Google Scholar] [CrossRef]
- Stephan, F. Phase shifts of circadian rhythms in activity entrained to food access. Physiol. Behav. 1984, 32, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Stephan, F.K. Resetting of a feeding-entrainable circadian clock in the rat. Physiol. Behav. 1992, 52, 985–995. [Google Scholar] [CrossRef]
- Davidson, A.J.; Aragona, B.J.; Werner, R.M.; Schroeder, E.; Smith, J.C.; Stephan, F.K. Food-anticipatory activity persists after olfactory bulb ablation in the rat. Physiol. Behav. 2001, 72, 231–235. [Google Scholar] [CrossRef]
- Damiola, F.; Minh, N.L.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schütz, G.; Schibler, U. Resetting of Circadian Time in Peripheral Tissues by Glucocorticoid Signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef]
- Davidson, A.J.; Castanon-Cervantes, O.; Leise, T.L.; Molyneux, P.C.; Harrington, M.E. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur. J. Neurosci. 2009, 29, 171–180. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Isejima, H.; Matsuo, T.; Okura, R.; Yagita, K.; Kobayashi, M.; Okamura, H. Synchronization of Cellular Clocks in the Suprachiasmatic Nucleus. Science 2003, 302, 1408–1412. [Google Scholar] [CrossRef]
- Nagano, M.; Adachi, A.; Nakahama, K.-I.; Nakamura, T.; Tamada, M.; Meyer-Bernstein, E.; Sehgal, A.; Shigeyoshi, Y. An Abrupt Shift in the Day/Night Cycle Causes Desynchrony in the Mammalian Circadian Center. J. Neurosci. 2003, 23, 6141–6151. [Google Scholar] [CrossRef]
- Davidson, A.J.; Poole, A.S.; Yamazaki, S.; Menaker, M. Is the food-entrainable circadian oscillator in the digestive system? Genes Brain Behav. 2003, 2, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Oda, G.A.; Niswender, K.D.; Yamazaki, S. Period determination in the food-entrainable and methamphetamine-sensitive circadian oscillator(s). Proc. Natl. Acad. Sci. USA 2012, 109, 14218–14223. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Yamazaki, S. The Mysterious Food-Entrainable Oscillator: Insights from Mutant and Engineered Mouse Models. J. Biol. Rhythm. 2018, 33, 458–474. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Yamazaki, S. Effects of light, food, and methamphetamine on the circadian activity rhythm in mice. Physiol. Behav. 2014, 128, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Wendroth, R.H.; Stenner, R.C.; Keil, C.D.; Yamazaki, S. mPeriod2 Brdm1 and other single Period mutant mice have normal food anticipatory activity. Sci. Rep. 2017, 7, 15510. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Nishijima, I.; Nakamura, W.; Nakamura, T.J. Secretin receptor-deficient mice exhibit robust food anticipatory activity. Neurosci. Lett. 2022, 772, 136462. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Nakamura, W.; Friday, R.C.; Hatanaka, F.; Takumi, T.; Yamazaki, S. Robust Food Anticipatory Activity in BMAL1-Deficient Mice. PLoS ONE 2009, 4, e4860. [Google Scholar] [CrossRef]
- Takasu, N.N.; Kurosawa, G.; Tokuda, I.T.; Mochizuki, A.; Todo, T.; Nakamura, W. Circadian Regulation of Food-Anticipatory Activity in Molecular Clock–Deficient Mice. PLoS ONE 2012, 7, e48892. [Google Scholar] [CrossRef]






| Tissue | RF for 2 Days | RF for 7 Days |
|---|---|---|
| SCN | 3.1 h (Phase Delay) | −1.0 h (Phase Advance) |
| Esophagus | −6.8 h (Phase Advance) | −1.7 h (Phase Advance) |
| Jejunum | 5.6 h (Phase Delay) | 7.0 h (Phase Delay) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakazawa, K.; Matsuo, M.; Kimura, N.; Numano, R. Restricted Feeding Resets the Peripheral Clocks of the Digestive System. Biomedicines 2023, 11, 1463. https://doi.org/10.3390/biomedicines11051463
Nakazawa K, Matsuo M, Kimura N, Numano R. Restricted Feeding Resets the Peripheral Clocks of the Digestive System. Biomedicines. 2023; 11(5):1463. https://doi.org/10.3390/biomedicines11051463
Chicago/Turabian StyleNakazawa, Kazuo, Minako Matsuo, Naobumi Kimura, and Rika Numano. 2023. "Restricted Feeding Resets the Peripheral Clocks of the Digestive System" Biomedicines 11, no. 5: 1463. https://doi.org/10.3390/biomedicines11051463