Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants
Abstract
:1. Introduction
2. Structural Characteristics of WRKY TFs
3. Regulatory Mechanism of WRKY TFs
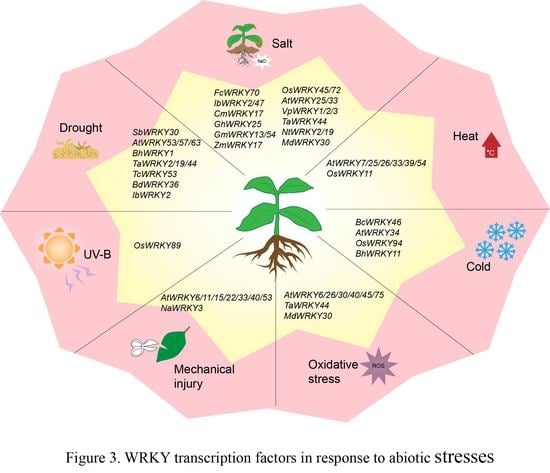
4. WRKY TF Involved in Abiotic Stress Responses
4.1. WRKY TFs and Drought Stress
4.2. WRKY TFs and Salt Stress
4.3. WRKY TFs and Temperature Stress
4.4. WRKY TFs and Other Abiotic Stresses
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yang, T.; Lin, Z.; Guo, B.; Xing, C.; Zhao, L.; Dong, H.; Gao, J.; Xie, Z.; Zhang, S.-L.; et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 2019, 17, 1770–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrestha, A.; Khan, A.; Dey, N. cis-trans Engineering: Advances and perspectives on customized transcriptional regulation in plants. Mol. Plant 2018, 11, 886–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Watanabe, S.; Tateno, M.; Seki, M.; Shinozaki, K.; Yokoyama, S. Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol. Biochem. 2008, 46, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.A.; Shapiro, H.; Nishiyama, T.; Perroud, P.-F.; Lindquist, E.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2007, 319, 64–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangelsen, E.; Kilian, J.; Berendzen, K.W.; Kolukisaoglu, Ü.; Harter, K.; Jansson, C.; Wanke, D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008, 9, 194. [Google Scholar] [CrossRef] [Green Version]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Berri, S.; Abbruscato, P.; Faivre-Rampant, O.; Brasileiro, A.C.M.; Fumasoni, I.; Satoh, K.; Kikuchi, S.; Mizzi, L.; Morandini, P.; Pè, M.E.; et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009, 9, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-J.; Ekramoddoullah, A.K. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome 2009, 52, 77–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, E.I. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [Green Version]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Zhang, Z.-L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2004, 137, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Bokowiec, M.T.; Han, S.; Zhang, H.; Brannock, J.F.; Chen, X.; Laudeman, T.W.; Timko, M.P. Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 2008, 147, 280–295. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta BBA Bioenerg. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef] [Green Version]
- Ezentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef] [Green Version]
- Turck, F.; Zhou, A.; Somssich, I.E. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley. Plant Cell 2004, 16, 2573–2585. [Google Scholar] [CrossRef] [Green Version]
- Cai, T.; Flanagan, L.B.; Jassal, R.S.; Black, T.A. Modelling environmental controls on ecosystem photosynthesis and the carbon isotope composition of ecosystem-respired CO2 in a coastal Douglas-fir forest. Plant Cell Environ. 2008, 31, 435–453. [Google Scholar] [CrossRef]
- Grierson, C.; Du, J.-S.; Zabala, M.D.T.; Beggs, K.; Smith, C.; Holdsworth, M.; Bevan, M.W. Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J. 1994, 5, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2008, 69, 91–105. [Google Scholar] [CrossRef]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.-D.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef]
- Zou, C.; Jiang, W.; Yu, D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 2010, 61, 3901–3914. [Google Scholar] [CrossRef] [Green Version]
- Park, C.Y.; Lee, J.H.; Yoo, J.H.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; Lee, S.M.; Kim, H.S.; Kang, K.Y.; Chung, W.S.; et al. WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 2005, 579, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015, 34, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ge, J.; Bao, C.; Chang, W.; Liu, J.; Shao, J.; Liu, X.; Su, L.; Pan, L.; Zhou, D.-X. Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 2020, 13, 598–611. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yu, D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.-K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ma, K.-B.; Lu, Z.-G.; Ren, S.-X.; Jiang, H.-R.; Cui, J.-W.; Chen, G.; Teng, N.-J.; Lam, H.-M.; Jin, B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Ren, S.; Ma, K.; Lu, Z.; Chen, G.; Cui, J.; Tong, P.; Wang, L.; Teng, N.; Jin, B. Transcriptomic and metabolomic analysis of the heat-stress response of Populus tomentosa. Carr. For. 2019, 10, 383. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, L.; Yan, Y.; Zhang, S.; Li, H.; Gao, Z.; Wang, C.; Guo, X. GhWRKY21 regulates ABA-mediated drought tolerance by fine-tuning the expression of GhHAB in cotton. Plant Cell Rep. 2020, 39. [Google Scholar] [CrossRef]
- Liu, X.; Song, Y.; Xing, F.; Wang, N.; Wen, F.; Zhu, C. GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 2015, 253, 1265–1281. [Google Scholar] [CrossRef]
- Jia, H.; Wang, C.; Wang, F.; Liu, S.; Li, G.; Guo, X. GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0120646. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Vannozzi, A.; Wang, G.; Liang, Y.-H.; Tornielli, G.B.; Zenoni, S.; Cavallini, E.; Pezzotti, M.; Cheng, Z.-M. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 2014, 1, 14016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef]
- Sun, J.; Hu, W.; Zhou, R.; Wang, L.; Wang, X.; Wang, Q.; Feng, Z.-J.; Yu, H.; Qiu, D.; He, G.; et al. The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 2014, 34, 23–35. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, J.; Hu, J.; Wang, W.; Wu, H.; Zhang, Q.; Liu, J.-H. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015, 38, 2248–2262. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, L.; Zhang, L.; Yu, D. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 2010, 35, 459–471. [Google Scholar] [CrossRef]
- Dai, X.; Wang, Y.; Zhang, W.-H. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 2016, 67, 947–960. [Google Scholar] [CrossRef] [Green Version]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef]
- Zhou, Q.-Y.; Tian, A.-G.; Zou, H.-F.; Xie, Z.-M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.-S.; Chen, S.-Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef]
- Niu, C.-F.; Wei, W.; Zhou, Q.-Y.; Tian, A.-G.; Hao, Y.-J.; Zhang, W.; Ma, B.; Lin, Q.; Zhang, Z.-B.; Zhang, J.-S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Wang, F.; Hou, X.; Tang, J.; Wang, Z.; Wang, S.; Jiang, F.; Li, Y. A novel cold-inducible gene from Pak-choi (Brassica campestris ssp. chinensis), BcWRKY46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco. Mol. Biol. Rep. 2011, 39, 4553–4564. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, Y.; Wang, L.; Liu, X.; Liu, Y.; Phillips, J.; Deng, X. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 2009, 230, 1155–1166. [Google Scholar] [CrossRef]
- Li, H.; Xu, Y.; Xiao, Y.; Zhu, Z.; Xie, X.; Zhao, H.; Wang, Y. Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 2010, 232, 1325–1337. [Google Scholar] [CrossRef]
- Zhu, Z.; Shi, J.; Cao, J.; He, M.; Wang, Y. VpWRKY3, a biotic and abiotic stress-related transcription factor from the Chinese wild Vitis pseudoreticulata. Plant Cell Rep. 2012, 31, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhang, Y.; Han, L.; Guan, Z.; Chai, T. A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep. 2008, 27, 795–803. [Google Scholar] [CrossRef]
- Skibbe, M.; Qu, N.; Galis, I.; Baldwin, I.T. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 2008, 20, 1984–2000. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Zhang, W.; Liu, Z.; Yi-Maer, A.Y.; Zhai, M.; Xu, Z. Both JrWRKY2 and JrWRKY7 of Juglans regia mediate responses to abiotic stresses and abscisic acid through formation of homodimers and interaction. Plant Biol. 2017, 19, 268–278. [Google Scholar] [CrossRef]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J.; et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 2020, 246, 153142. [Google Scholar] [CrossRef]
- Song, Y.; Li, J.; Sui, Y.; Han, G.; Zhang, Y.; Guo, S.; Sui, N. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 2020, 102, 603–614. [Google Scholar] [CrossRef]
- Qin, Z.; Hou, F.; Li, A.; Dong, S.; Wang, Q.; Zhang, L. Transcriptome-wide identification of WRKY transcription factor and their expression profiles under salt stress in sweetpotato (Ipomoea batatas L.). Plant Biotechnol. Rep. 2020, 14, 599–611. [Google Scholar] [CrossRef]
- Zhu, H.; Zhou, Y.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic. Arabidopsis 2020, 10, 506. [Google Scholar] [CrossRef] [Green Version]
- Dong, Q.; Zheng, W.; Duan, D.; Huang, D.; Wang, Q.; Liu, C.; Li, C.; Gong, X.; Li, C.; Mao, K.; et al. MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 2020, 299, 110611. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2020, 18. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Li, X.; Yang, Y.; Liu, C.; Zhou, G.; Wan, H.; Cheng, Y. Tomato WRKY81 acts as a negative regulator for drought tolerance by modulating guard cell H2O2–mediated stomatal closure. Environ. Exp. Bot. 2020, 171, 103960. [Google Scholar] [CrossRef]
- Luo, X.; Li, C.; He, X.; Zhang, X.; Zhu, L.-F. ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 2019, 39, 181–194. [Google Scholar] [CrossRef]
- Wang, M.-Q.; Huang, Q.-X.; Lin, P.; Zeng, Q.-H.; Li, Y.; Liu, Q.-L.; Zhang, L.; Pan, Y.-Z.; Jiang, B.-B.; Zhang, F. The overexpression of a transcription factor gene VbWRKY32 enhances the cold tolerance in Verbena bonariensis. Front. Plant Sci. 2020, 10, 1746. [Google Scholar] [CrossRef]
- Chanwala, J.; Satpati, S.; Dixit, A.; Parida, A.; Giri, M.K.; Dey, N. Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom. 2020, 21, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Zhang, D.; Lv, K.; Zhang, X.; Cheng, Z.; Li, R.; Zhou, B.; Jiang, T. Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci. 2019, 289, 110259. [Google Scholar] [CrossRef]
- Anjum, S.A.; Xie, X.-Y.; Wang, L.-C.; Saleem, M.F.; Man, C.; Lei, W. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agr. Res. 2011, 6, 2026–2032. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, S.; Lethin, J.; Blomberg, R.; Mousavi, H.; Aronsson, H. In silico based screening of WRKY genes for identifying functional genes regulated by WRKY under salt stress. Comput. Biol. Chem. 2019, 83, 107131. [Google Scholar] [CrossRef]
- Linxiao, W.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef]
- Shen, Z.; Yao, J.; Sun, J.; Chang, L.; Wang, S.; Ding, M.; Qian, Z.; Zhang, H.; Zhao, N.; Sa, G.; et al. Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Sci. 2015, 235, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhao, Y.; Xu, S.; Zhang, Z.; Xu, Y.; Zhang, J.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Jin, B.; Wang, L.; Wang, J.; Jiang, K.-Z.; Wang, Y.; Jiang, X.X.; Ni, C.-Y.; Wang, Y.; Teng, N.-J. The effect of experimental warming on leaf functional traits, leaf structure and leaf biochemistry in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.-T.; Li, G.-L.; Chang, H.; Sun, D.-Y.; Zhou, R.-G.; Li, B. Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 2007, 30, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Huang, B. Effects of abscisic acid, salicylic acid, ethylene and hydrogen peroxide in thermotolerance and recovery for creeping bentgrass. Plant Growth Regul. 2005, 47, 17–28. [Google Scholar] [CrossRef]
- Zhao, J.; He, Q.; Chen, G.; Wang, L.; Jin, B. Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 2016, 7, 1213. [Google Scholar] [CrossRef] [Green Version]
- Gong, X.-Q.; Hu, J.-B.; Liu, J.-H. Cloning and characterization of FcWRKY40, A WRKY transcription factor from Fortunella crassifolia linked to oxidative stress tolerance. Plant Cell Tissue Organ Cult. PCTOC 2014, 119, 197–210. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Chang, H.-S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002, 129, 661–677. [Google Scholar] [CrossRef] [Green Version]
- Robatzek, S.; Somssich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001, 28, 123–133. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, X.; Wang, Y.; Ming, F. The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 2013, 8, e73541. [Google Scholar] [CrossRef] [PubMed]



| No. | Gene | Species | Induced by Factors | Function | References |
|---|---|---|---|---|---|
| 1 | AtWRKY25/26 | Arabidopsis | Heat | Tolerance to heat | [34] |
| 2 | AtWRKY33 | Arabidopsis | NaCl, mannitol, H2O2 | Tolerance to heat and NaCl, negative regulator in oxidative stress and abscisic acid (ABA) | [33] |
| 3 | AtWRKY34 | Arabidopsis | Cold | Negative regulator in cold stress | [35] |
| 4 | AtWRKY39 | Arabidopsis | Heat | Tolerance to heat | [36] |
| 5 | AtWRKY53 | Arabidopsis | Drought, salt | Reduced drought resistance and H2O2, sensitive to salt | [37,38] |
| 6 | AtWRKY57 | Arabidopsis | Drought | Tolerance to drought | [39] |
| 7 | AtWRKY63 | Arabidopsis | ABA | Tolerance to drought, regulated ABA signaling | [40] |
| 8 | AtWRKY54 | Arabidopsis | Heat | Response to heat stress | [41] |
| 9 | POWRKY13 | Populus tomentosa | Heat | Response to heat stress | [42] |
| 10 | GhWRKY21 | Gossypium hirsutum | Drought | Tolerance to drought | [43] |
| 11 | GhWRKY25 | Gossypium hirsutum | Drought | Tolerance to salt, reduced drought resistance | [44] |
| 12 | GhWRKY68 | Gossypium hirsutum | Salt, drought | Reduced salt tolerance and drought resistance, positive regulator in ABA signaling | [45] |
| 13 | VvWRKY24 | Vitis vinifera | Cold | Upregulated expression at all stages of hypothermia | [46] |
| 14 | CaWRKY40 | Capsicum annuum | Heat | Tolerance to heat | [47] |
| 15 | BdWRKY36 | Brachypodium distachyon | Drought | Tolerance to drought | [48] |
| 16 | FcWRKY70 | Fortunella crassifolia | Salt | Tolerance to salt | [49] |
| 17 | OsWRKY11 | Oryza sativa | Heat, drought | Tolerance to drought and heat | [50] |
| 18 | OsWRKY72 | Oryza sativa | Drought, NaCl, ABA | Sensitive to salt, drought, sucrose, and ABA | [51] |
| 19 | OsWRKY74 | Oryza sativa | Pi deprivation, cold | Tolerance to cold and Pi deprivation | [52] |
| 20 | OsWRKY76 | Oryza sativa | Cold | Tolerance to cold | [53] |
| 21 | OsWRKY89 | Oryza sativa | ABA, UV-B | Tolerance to UV | [54] |
| 22 | GmWRKY13 | Soybean | Salt, drought | Sensitive to salt and mannitol, negative regulator in ABA signaling | [55] |
| 23 | GmWRKY17 | Soybean | Salt | Reduced salt tolerance | [56] |
| 24 | GmWRKY54 | Soybean | Salt, drought | Tolerance to salt and drought | [55] |
| 25 | GmWRKY21 | Glycine max | NaCl, drought, cold | Tolerance to cold | [55] |
| 26 | ZmWRKY17 | Zea mays | ABA, salt | Reduced salt tolerance | [57] |
| 27 | TaWRKY2/19 | Triticum aestivum | NaCl, drought, ABA | Tolerance to salt and drought | [58] |
| 28 | BcWRKY46 | Brassica campestris | NaCl, drought, cold | Tolerance to salt and drought | [59] |
| 29 | BhWRKY1 | Boea hygrometrica | Dehydration, ABA | Tolerance to drought | [60] |
| 30 | VpWRKY1 | Vitis pseudoreticulata | NaCl, ABA | Tolerance to salt | [61] |
| 31 | VpWRKY2 | Vitis pseudoreticulata | Cold, NaCl, ABA | Tolerance to salt and cold | [61] |
| 32 | VpWRKY3 | Vitis pseudoreticulata | Drought, ABA, salicylic acid (SA) | Tolerance to salt | [62] |
| 33 | TcWRKY53 | Thlaspi caerulescens | Cold, PEG, NaCl | Negative regulator in osmotic stress | [63] |
| 34 | NaWRKY3 | Nicotiana attenuate | Mechanical damage | Sensitive to mechanical damage | [64] |
| 35 | JrWRKY2/7 | Juglans regia | Drought, cold | Tolerance to drought and cold | [65] |
| 36 | SbWRKY30 | Sorghum bicolor | Salt, drought | Tolerance to salt and drought | [66] |
| 37 | SbWRKY50 | Sorghum bicolor | Salt | Tolerance to salt | [67] |
| 38 | IbWRKY47 | Ipomoea batatas | Salt | Tolerance to salt | [68] |
| 39 | IbWRKY2 | Ipomoea batatas | Salt, drought | Tolerance to salt and drought | [69] |
| 40 | MdWRKY30 | Malus domestica | Salt, osmotic stress | Tolerance to salt and osmotic stress | [70] |
| 41 | MdWRKY100 | Malus domestica | Salt | Sensitive to salt | [71] |
| 42 | SlWRKY81 | Solanum lycopersicum | Drought | Reduced drought tolerance | [72] |
| 43 | GbWRKY1 | Gossypium barbadense | Salt | Tolerance to salt | [73] |
| 44 | VbWRKY32 | Verbena bonariensis | Cold | Tolerance to cold | [74] |
| 45 | PgWRKY33/62 | Pennisetum glaucum | Salt, drought | Tolerance to salt and drought | [75] |
| 46 | PagWRKY75 | Populus alba | Drought | Negative regulator in salt and osmotic stress | [76] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. https://doi.org/10.3390/plants9111515
Li W, Pang S, Lu Z, Jin B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants. 2020; 9(11):1515. https://doi.org/10.3390/plants9111515
Chicago/Turabian StyleLi, Weixing, Siyu Pang, Zhaogeng Lu, and Biao Jin. 2020. "Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants" Plants 9, no. 11: 1515. https://doi.org/10.3390/plants9111515