Evolution and Classification of Musaceae Based on Male Floral Morphology
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis
2.2. Multivariate Analysis
2.3. Morphological Analysis
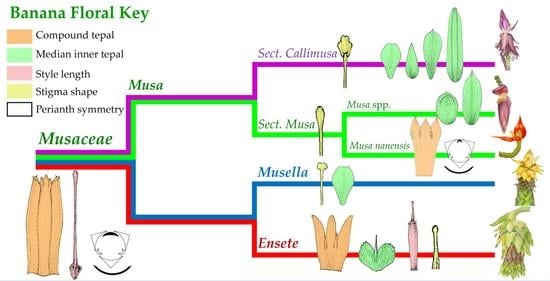
2.4. Morphological Characters in the Classification of Musaceae
3. Discussion
3.1. Identification of Musa Species Using Floral Morphological Characters
3.2. Taxonomic Treatment of Musaceae Based on Floral Morphology
| Key to the genera of Musaceae |
| 1a. Outer tepal lobe apex round, lateral inner tepal acicular, style short (less than 18 mm length) ………………………………………………………………………………………… Ensete |
| 1b. Outer tepal lobe apex acute, lateral inner tepal ovate in shape, style long (more than 18 mm length) …………….…………………………………………………………………………… 2 |
| 2a. Stigma clavate or spatulate ………………..……………………………………………. Musa |
| 2b. Stigma capitate …………..………………………………………………… Musella lasiocarpa |
| Key to species of Ensete |
| 1a. Length/width ratio of fused tepal less than 3:1, base of median inner tepal ob- tuse……………………………………………………………………………………...... E. homblei |
| 1b. Length/width ratio of fused tepal more than 3:1, base of median tepal acute or subcor- date…………………………………………………………………………………………………. 2 |
| 2a. Base of median inner tepal acute, margin of median inner tepal shoulder den- tate……………………………………………………………………………………….... E. glaucum |
| 2b. Base of median inner tepal subcordate, margin of median inner tepal shoulder entire..3 |
| 3a. Shoulder of median inner tepal round ………………………………….…………. E. gilletii |
| 3b. Shoulder of median inner tepal acute………………………………………...... E. superbum |
| Key to the section of Musa |
| 1a. Median inner tepal ovate or elliptic in shape, or missing, stigma clavate …………..………………………………………………….. sect. Musa (including Rhodochlamys |
| 1b. Median inner tepal oval-lanceolate, oblanceolate, oblong or obovate, stigma spatulate ……………………………………….... sect. Callimusa (including Australimusa & Ingentimusa) |
| Key to species of the section Musa |
| 1a. Median inner tepal ovate, shoulder margin repand ……………………………………… 2 |
| 1b. Median inner tepal elliptic, shoulder margin dentate or entire, or fused with lateral outer tepal ………………………………………………………………………………………….. 3 |
| 2a. Perianth ventricose, anther ensiform …………………………………………. M. nagensium |
| 2b. Perianth tubular, anther oblong……………………………………………………………… 4 |
| 3a. Flowers nearly actinomorphic; median inner tepal fused with lateral outer tepals at adaxial side; fertile stamens six, filaments basally united, anther ensiform…… M. nanensis |
| 3b. Flower zygomorphic; median inner tepal elliptic and free; fertile stamens five, free, an- ther oblong………………………………………………………………………………………….. 5 |
| 4a. Wart-like structures on styles near stigma absent ……………………………… M. velutina |
| 4b. Wart-like structures on styles near stigma present ………………………………. M. ornata |
| 5a. Wrinkle on median inner tepal absent; surface of stigma velvet……………. M. balbisiana |
| 5b. Wrinkle on median inner tepal present; surface of stigma smooth …………………….. 6 |
| 6a. Perianth tubular; length/width ratio of fused tepal less than 3:1…. M. rubra, M. siamensis |
| 6b. Perianth ventricose; length/width ratio of fused tepal more than 3:1 ………………….. 7 |
| 7a. Spine-like dorsal appendage on lateral inner tepal lobes present ……………M. itinerans |
| 7b. Spine-like dorsal appendage on lateral inner tepal lobes absent ……………………….. 8 |
| 8a. Apex of median inner tepal truncate……………………………………………. M. serpentina |
| 8b. Apex of median inner tepal acute ………………………………………………………....... 9 |
| 9a. Median inner tepal base truncate, wing on median inner tepals present………………... …………………………………………………………………………………….….M. yunnanensis |
| 9b. Median inner tepal base obtuse, wing on median inner tepals absent…………………... ……………………………………………………………………………M. acuminata, M. flaviflora |
| Key to the species of the section Callimusa |
| 1a. Fused tepal length to median inner tepal length ratio 1–2 …………….………………… 2 |
| 1b. Fused tepal length to median inner tepal length ratio 3–4 ………………………………. 3 |
| 2a. Spine-like dorsal appendage on median outer tepal lobes absent, median inner tepal apex acuminate, wing on shoulder of median inner tepal present and margin on shoulder of median inner tepal dentate ………………………………………………………... M. gracilis |
| 2b. Spine-like dorsal appendage on median outer tepal lobes present, median inner tepal apex subobtuse, wing on shoulder of median inner tepal absent and margin on shoulder of median inner tepal entire ………………………………………………………….………… 4 |
| 3a. Median inner tepal obovate, margin of median inner tepal shoulder dentate, shoulder of median inner tepal acute, wrinkle on median inner tepal present ….………... M. textilis |
| 3b. Median inner tepal oval-lanceolate or oblanceolate, margin of median inner tepal shoulder entire, shoulder of median inner tepal round, wrinkle on median inner tepal ab- sent ………………………………………………………………………………………………… 5 |
| 4a. Spine-like dorsal appendage on lateral inner tepal lobes present, shoulder of median inner tepal round and ovary transverse section angular…………………… M. paracoccinea |
| 4b. Spine-like dorsal appendage on lateral inner tepal lobes absent, shoulder of median inner tepal truncate and ovary transverse section cylindrical ……………………………... 6 |
| 5a. Median inner tepal oval-lanceolate and median inner tepal base obtuse……M. beccarii |
| 5b. Median inner tepal oblanceolate and median inner tepal base acute ….……..…………… ………………………………………………………………………………… M. maclayi, M. ingens |
| 6a. Spine-like dorsal appendage on lateral outer tepal lobes absent…………... M. haekkinenii |
| 6b. Spine-like dorsal appendage on lateral outer tepal lobes present…………..… M. coccinea |
4. Materials and Methods
4.1. Taxon Sampling
4.2. Molecular Methods
4.3. Sequence Alignment and Phylogeny Reconstruction
4.4. Multivariate Analysis
4.5. Morphological Analyses
4.6. Character Evolution and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kress, W.J. The phylogeny and classification of the Zingiberales. Ann. Mo. Bot. Gard. 1990, 77, 698–721. [Google Scholar] [CrossRef]
- Cheesman, E.E. Classification of the bananas. I. The genus Ensete Horan. Kew Bull. 1947, 2, 97–106. [Google Scholar] [CrossRef]
- Argent, G. The wild bananas of Papua New Guinea. Notes Roy. Bot. Gard. Edinburgh. 1976, 35, 77–114. [Google Scholar]
- Hakkinen, M. Reappraisal of section taxonomy in Musa (Musaceae). Taxon 2013, 62, 809–813. [Google Scholar] [CrossRef] [Green Version]
- Swangpol, S.C.; Traiperm, P.; Somana, J.; Sukkaewmanee, N.; Srisanga, P.; Suksathan, P. Musa nanensis, a new banana (Musaceae) species from Northern Thailand. Syst. Bot. 2015, 40, 426–432. [Google Scholar] [CrossRef]
- Liu, A.Z.; Kress, W.J.; Li, D.Z. Phylogenetic analyses of the banana family (Musaceae) based on nuclear ribosomal (ITS) and chloroplast (trnL-F) evidence. Taxon 2010, 59, 20–28. [Google Scholar] [CrossRef]
- Li, L.F.; Häkkinen, M.; Yuan, Y.M.; Hao, G.; Ge, X.J. Molecular phylogeny and systematics of the banana family (Musaceae) inferred from multiple nuclear and chloroplast DNA fragments, with a special reference to the genus Musa. Mol. Phylogenet. Evol. 2010, 57, 1–10. [Google Scholar] [CrossRef]
- Janssens, S.B.; Vandelook, F.; De Langhe, E.; Verstraete, B.; Smets, E.; Vandenhouwe, I.; Swennen, R. Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol. 2016, 210, 1453–1465. [Google Scholar] [CrossRef] [Green Version]
- Rudall, P.J.; Bateman, R.M. Evolution of zygomorphy in monocot flowers: Iterative patterns and developmental constraints. New Phytol. 2004, 162, 25–44. [Google Scholar] [CrossRef]
- Bartlett, M.E.; Specht, C.D. Evidence for the involvement of GLOBOSA -like gene duplications and expression divergence in the evolution of floral morphology in the Zingiberales. New Phytol. 2010, 187, 521–541. [Google Scholar] [CrossRef]
- Chessman, E.E. Classification of the Bananas. Kew Bull. 1949, 4, 450–452. [Google Scholar] [CrossRef]
- Hooker, J.D. Musa hillii. In Curtis’s Botanical Magazine. V. 121; L. Reeve & Co.: London, UK, 1895; p. 7401. [Google Scholar]
- Cheesman, E.E. Classification of the Bananas. Kew Bull. 1950, 5, 27–28. [Google Scholar] [CrossRef]
- Argent, G. Two interesting wild Musa species (Musaceae) from Sabah, Malaysia. Gard. Bull. Singapore 2000, 52, 203–210. Available online: https://www.biodiversitylibrary.org/part/122250 (accessed on 16 March 2023).
- Häkkinen, M.; Meekiong, K. Musa borneensis Becc. (Musaceae) and its intraspecific taxa in Borneo. Acta Phytotax.GeoboL. 2005, 56, 213–230. Available online: https://www.musalit.org/seeMore.php?id=10981 (accessed on 16 March 2023).
- Häkkinen, M. Musa barioensis, New Musa species (Musaceae) from Northern Borneo. Acta Phytotax.GeoboL. 2006, 57, 55–60. Available online: https://www.jstage.jst.go.jp/article/apg/57/1/57_KJ00004622843/_article (accessed on 16 March 2023).
- Häkkinen, M. Musa campestris Becc. (Musaceae) varieties in northern Borneo. Folia Malays. 2004, 5, 81–100. Available online: https://www.musalit.org/seeMore.php?id=8671 (accessed on 16 March 2023).
- Valmayor, R.V. Classification et caractérisation de Musa exotica, M. alinsanaya et M. acuminata ssp. Errans [Classification and characterization of Musa exotica, M. alinsanaya and M. acuminata ssp. errans]. Infomusa 2001, 10, 35–39. [Google Scholar]
- Beccari, O. Nota sui banani selvatici di Borneo. In Nelle foreste di Borneo; Tipografia di Salvadore Landi: Firenze, Italy, 1902; p. 624. [Google Scholar]
- Cheesman, E.E. Classification of the Bananas: Critical Notes on Species: M. violascens and M. gracilis. Kew Bull. 1950, 5, 152–155. [Google Scholar] [CrossRef]
- Häkkinen, M.; Vare, H. Typification of Musa salaccensis and nomenclatural notes on Musa (Musaceae). Adansonia 2009, 31, 41–46. [Google Scholar] [CrossRef]
- Valmayor, R.V.; Danh, L.D.; Hakkinen, M. Rediscovery of Musa splendida A. Chevalier and description of two new species (Musa viridis and Musa lutea). Philipp. Agric. Sci. 2004, 87, 110–118. [Google Scholar]
- Häkkinen, M.; Tea, C.H. Musa rubinea, a new Musa species (Musaceae) from Yunnan, China. Folia Malays. 2008, 9, 23–33. [Google Scholar]
- Joe, A.; Sreejith, P.E.; Sabu, M. Notes on the rediscovery, taxonomic history and conservation of Musa mannii H. Wendl. ex Baker (Musaceae). Webbia 2014, 69, 117–122. [Google Scholar] [CrossRef]
- Häkkinen, M.; Vare, H. A taxonomic revision of Musa aurantiaca (Musaceae) in Southeast Asia. J. Syst. Evol. 2008, 46, 89–92. [Google Scholar]
- Baker, J.G. A synopsis of the genera and species of Museae. Ann. Bot. 1893, 7, 189–229. [Google Scholar] [CrossRef]
- Simmonds, N.W.; Willison, N. Botanical Results of the Banana Collecting Expedition, 1954-5. Kew Bull. 1956, 11, 463–489. [Google Scholar] [CrossRef]
- Valmayor, R.V.; Danh, L.D.; Häkkinen, M. The wild and ornamental Musaceae of Vietnam with descriptions of two new traveling bananas. Philipp. Agric. Sci. 2005, 88, 236–244. Available online: https://www.musalit.org/seeMore.php?id=9375 (accessed on 16 March 2023).
- Joe, A.; Sreejith, P.E.; Sabu, M. On the Discovery and extended distribution of Musa cheesmanii Musaceae from North-East India. J. Plant Anim. Environ. Sci. 2014, 4, 1–4. [Google Scholar]
- Simmonds, N.W. Notes on banana taxonomy. Kew Bull. 1960, 14, 198–212. [Google Scholar] [CrossRef]
- Bekele, E.; Shigeta, M. Phylogenetic relationships between Ensete and Musa species as revealed by the trnT trnF region of cpDNA. Genet. Resour. Crop Evol. 2011, 58, 259–269. [Google Scholar] [CrossRef]
- Christelova, P.L.; Valarik, M.R.; Hribova, E.V.; De Langhe, E.; Dolezel, J.R. A multi gene sequence-based phylogeny of the Musaceae (banana) family. BMC Evol. Biol. 2011, 11, 103. Available online: http://www.biomedcentral.com/1471-2148/11/103 (accessed on 16 March 2023). [CrossRef] [Green Version]
- Wu, D.L.; Kress, W.J. Musaceae. In Flora of China; Wu, C.Y., Raven, P.H., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2001; Volume 24, pp. 314–318. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.Z. Phylogeny and Biogeography of Musaceae. Dissertation, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China, 2001. [Google Scholar]
- Liu, A.Z.; Li, D.Z.; Li, X.W. Taxonomic notes on wild bananas (Musa) from China. Bot. Bull. Acad. Sin. 2002, 43, 77–81. [Google Scholar]
- Xue, C.Y.; Wang, H.; Li, D.Z. Female gametophyte and seed development in Musella lasiocarpa (Musaceae), a monotypic genus endemic to Southwestern China. Canad. J. Bot. 2007, 85, 964–975. [Google Scholar] [CrossRef]
- Cheesman, E.E. Classification of bananas II. The genus Musa L. Kew Bull. 1947, 2, 106–117. [Google Scholar] [CrossRef]
- Liu, A.-Z.; Kress, W.J.; Wang, H.; Li, D.-Z. Insect pollination of Musella lasiocarpa (Musaceae), a monotypic genus endemic to Yunnan, China. Pl. Syst. Evol. 2002, 235, 135–146. [Google Scholar] [CrossRef]
- Specht, C.D.; Yockteng, R.; Almeida, A.M.; Kirchoff, B.K.; Kress, W.J. Homoplasy, Pollination, and Emerging Complexity During the Evolution of Floral Development in the Tropical Gingers (Zingiberales). Bot. Rev. 2012, 78, 440–462. [Google Scholar] [CrossRef]
- Nur, N. Studies on pollination in Musaceae. Ann. Bot. 1975, 40, 167–177. [Google Scholar] [CrossRef]
- Ren, Z.; Wang1, H. Morphological comparison of floral nectaries in Musaceae, with reference to its pollinators. Biodivers. Sci. 2007, 15, 652–657. [Google Scholar] [CrossRef]
- Scott-Ellot, G.F. Note on the fertilization of Musa, Strelitzia reginae, and Ravenala madagascariensis. Ann. Bot. 1890, 4, 259–264. [Google Scholar] [CrossRef]
- Cronk, Q.; Ojeda, I. Bird-pollinated flowers in an evolutionary and molecular context. J. Exp. Bot. 2008, 59, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Endress, P.K. Diversity and Evolutionary Biology of Tropical Fowers; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Tomlinson, P.B. Phylogeny of the Scitamineae—Morphological and anatomical considerations. Evolution 1961, 16, 192–213. [Google Scholar] [CrossRef]
- Gawel, N.J.; Jarret, R.L. Chloroplast DNA restriction fragment length polymorphisms (RFLPs) in Musa species. Theor. Appl. Genet. 1991, 81, 783–786. [Google Scholar] [CrossRef]
- Ude, G.; Pillay, M.; Nwakanma, D.; Tenkouano, A. Analysis of genetic diversity and sectional relationships in Musa using AFLP markers. Theor. Appl. Genet. 2002, 104, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Kiew, R.; Agent, G.; Set, O.; Lee, S.K.; Gan, Y.Y. Assessment of validity of sections in Musa (Musaceae) using AFLP. Ann. Bot. 2002, 90, 231–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joe, A.; Sreejith, P.E.; Sabu, M. Notes on Musa rubra Kurz (Musaceae) and reduction of M. laterita Cheesman as conspecific. Taiwania 2016, 61, 34–40. [Google Scholar]
- Preston, J.C.; Hileman, L.C. Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. EvoDevo 2012, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citerne, H.; Jabbour, F.; Nadot, S.; Damerval, C. The evolution of floral symmetry. Adv. Bot. Res. 2010, 54, 87–137. [Google Scholar] [CrossRef]
- Hileman, L.C. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130348. [Google Scholar] [CrossRef] [Green Version]
- Voigt, J.O. Musaceae. In Hortus Suburbanus Calcuttensis; Bishop’s College Press: Calcutta, India, 1845; pp. 578–579. [Google Scholar]
- Hooker, J.D. Musa rubra. In Curtis’s Botanical Magazine. V. 121; L. Reeve & Co.: London, UK, 1895; p. 7451. [Google Scholar]
- Cheesman, E.E. Classification of the bananas III. The genus Musa L. Kew Bull. 1947, 3, 265–272. [Google Scholar] [CrossRef]
- Hakkinen, M.; Houng, W. New species and variety of Musa (Musaceae) from Yunnan, China. Novon 2007, 17, 440–446. [Google Scholar] [CrossRef]
- Inta, W.; Kongsawadworakul, P.; Viboonjun, U.; Chuenwarin, P.; Traiperm, P.; Sasivimon, S.C. Proposal to reduce anthocyanin-deficient banana Musa siamensis to a M. rubra variety. In Proceedings of the International Conference on Biodiversity, IBD2019, Bangkok, Thailand, 22–24 May 2019; pp. 116–121. [Google Scholar]
- Häkkinen, M.; Gogoi, R.; Borah, S. A taxonomic study of Musa flaviflora and M. thomsonii (Musaceae). Nord. J. Bot. 2014, 32, 578–583. [Google Scholar] [CrossRef]
- Joe, A.; Sreejith, P.E.; Sabu, M. Notes on the rediscovery and taxonomic status of M. flaviflora and M. thomsonii (Musaceae) from North-East India. Ann. Plant Sci. 2013, 2, 160–162. [Google Scholar]
- Doyle, J.J.; Dolye, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Oxelman, B.; Liden, M.; Berglund, D. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst. Evol. 1997, 206, 393–410. [Google Scholar] [CrossRef] [Green Version]
- Hurr, K.A.; Lockhart, P.J.; Heenan, P.B.; Penny, D. Evidence for the recent dispersal of Sophora (Leguminosae) around the southern oceans: Molecular data. J. Biogeogr. 1999, 26, 565–577. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Schwartz, T.; Pickett, B.E.; He, S.; Klem, E.B.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A. A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evol. Bioinform. Online 2015, 11, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougement, J. A fast bootstrapping algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. Available online: http://palaeo-electronica.org (accessed on 1 January 2023).
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis, Version 3.6. 2018. Available online: http://www.mesquiteproject.org/ (accessed on 26 September 2019).
- Fitch, W.M. Toward Defining Course of Evolution: Minimum Change for a Specific Tree Topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]










| Component | Initial Eigenvalues | ||
|---|---|---|---|
| Total | % of Variance | Cumulative % | |
| 1 | 5.289 | 35.26 | 35.26 |
| 2 | 3.980 | 26.53 | 61.79 |
| 3 | 1.496 | 9.98 | 71.76 |
| 4 | 1.312 | 8.75 | 80.51 |
| Component Matrix | Component | |||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 1. width of compound tepal | 0.730 | 0.345 | −0.199 | −0.297 |
| 2. length of compound tepal | 0.675 | 0.518 | 0.292 | 0.193 |
| 3. width of lateral outer compound tepal lobe | 0.845 | −0.267 | −0.296 | −0.173 |
| 4. length of lateral outer compound tepal lobe | 0.808 | −0.465 | −0.094 | −0.010 |
| 5. width of central outer compound tepal lobe | 0.861 | −0.332 | −0.161 | 0.008 |
| 6. length of central outer compound tepal lobe | 0.756 | −0.567 | −0.090 | 0.036 |
| 7. width of inner compound tepal lobe | 0.601 | 0.372 | −0.365 | 0.353 |
| 8. length of inner compound tepal lobe | 0.261 | −0.829 | 0.304 | −0.101 |
| 9. width of median inner tepal | −0.274 | 0.187 | 0.426 | 0.281 |
| 10. length of median inner tepal | 0.280 | 0.824 | 0.157 | −0.068 |
| 11. length of anther | 0.609 | −0.017 | 0.432 | 0.538 |
| 12. length of filament | 0.389 | 0.597 | 0.265 | −0.499 |
| 13. length from style base to stigma head | 0.475 | 0.733 | −0.105 | 0.375 |
| 14. width of ovary | 0.487 | 0.206 | 0.539 | −0.478 |
| 15. length of ovary | 0.227 | −0.630 | 0.502 | 0.169 |
| Eigenvalues | |||||
|---|---|---|---|---|---|
| Function | Eigenvalue | % of Variance | Cumulative % | Canonical Correlation | |
| 1 | 11.316 | 91.9 | 91.9 | 0.959 | |
| 2 | 0.996 | 8.1 | 100 | 0.706 | |
| Structure Matrix | Function | |
|---|---|---|
| 1 | 2 | |
| Length from style base to stigma head | 0.662 | −0.461 |
| Width of ovary | 0.252 | −0.064 |
| Length of median inner tepal | 0.231 | −0.031 |
| Length of filament | 0.181 | −0.169 |
| Length of ovary | −0.135 | 0.068 |
| Length of compound tepal | 0.200 | −0.683 |
| Length of anther | 0.024 | −0.487 |
| Width of lateral outer compound tepal lobe | −0.081 | −0.394 |
| Width of compound tepal | −0.069 | −0.393 |
| Length of inner compound tepal lobe | −0.109 | −0.389 |
| Width of central outer compound tepal lobe | −0.064 | −0.353 |
| Length of lateral outer compound tepal lobe | −0.083 | −0.241 |
| Width of inner compound tepal lobe | 0.001 | −0.195 |
| Length of central outer compound tepal lobe | −0.134 | −0.160 |
| Width of median inner tepal | 0.021 | 0.105 |
| No. | Male Floral Morphological Characters | Character States |
|---|---|---|
| 1 | Shape of perianth | (0) fused tepal with deep lobes |
| (1) ventricose | ||
| (2) tubular | ||
| 2 | Symmetry of perianth | (0) bilateral symmetry |
| (1) radial symmetry | ||
| 3 | Ratio of length/width of fused tepal | (0) 3–4 |
| (1) 1–2 | ||
| 4 | Shape of outer tepal apex lobe | (0) round |
| (1) acute | ||
| 5 | Spine-like dorsal appendage present on lateral outer tepal lobe | (0) absent |
| (1) present | ||
| 6 | Spine-like dorsal appendage present on median outer tepal lobe | (0) absent |
| (1) present | ||
| 7 | Shape of lateral inner tepal lobe | (0) acicular |
| (1) ovate | ||
| 8 | Spine-like dorsal appendage present on lateral inner tepal lobe | (0) absent |
| (1) present | ||
| 9 | Shape of median inner tepal | (0) tricuspidate |
| (1) obovate | ||
| (2) oval-lanceolate | ||
| (3) oblanceolate | ||
| (4) oblong | ||
| (5) elliptic | ||
| (6) ovate | ||
| 10 | Base of median inner tepal | (0) acute |
| (1) sub-cordate | ||
| (2) obtuse | ||
| (3) truncate | ||
| (4) adnate with fused tepal | ||
| 11 | Apex of free median inner tepal | (0) cuspidate |
| (1) acuminate | ||
| (2) attenuate | ||
| (3) subobtuse | ||
| 12 | Wings on shoulder of free median inner tepal | (0) present |
| (1) absent | ||
| 13 | Margin of free median inner tepal shoulder | (0) dentate |
| (1) entire | ||
| (2) repand | ||
| 14 | Shoulder of free median inner tepal | (0) acute |
| (1) round | ||
| (2) truncate | ||
| 15 | Present of wrinkle on free median inner tepal | (0) absent |
| (1) present | ||
| 16 | Ratio of median inner tepal length/fused tepal length | (0) 1/3–1/4 |
| (1) 1/1–1/2 | ||
| 17 | Shape of anther | (0) oblong |
| (1) ensiform | ||
| 18 | Shape of stigma | (0) capitate |
| (1) clavate | ||
| (2) spatulate | ||
| 19 | Surface of stigma | (0) smooth |
| (1) velvet | ||
| 20 | Wart-like structure present on style surface | (0) absent |
| (1) present | ||
| 21 | Ovary transvers shape | (0) cylindrical |
| (1) angular | ||
| 22 | Style length | (0) less than 18 mm |
| (1) more than 20 mm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inta, W.; Traiperm, P.; Ruchisansakun, S.; Janssens, S.B.; Viboonjun, U.; Swangpol, S.C. Evolution and Classification of Musaceae Based on Male Floral Morphology. Plants 2023, 12, 1602. https://doi.org/10.3390/plants12081602
Inta W, Traiperm P, Ruchisansakun S, Janssens SB, Viboonjun U, Swangpol SC. Evolution and Classification of Musaceae Based on Male Floral Morphology. Plants. 2023; 12(8):1602. https://doi.org/10.3390/plants12081602
Chicago/Turabian StyleInta, Wandee, Paweena Traiperm, Saroj Ruchisansakun, Steven B. Janssens, Unchera Viboonjun, and Sasivimon C. Swangpol. 2023. "Evolution and Classification of Musaceae Based on Male Floral Morphology" Plants 12, no. 8: 1602. https://doi.org/10.3390/plants12081602