Identification of High Tolerance to Jujube Witches’ Broom in Indian Jujube (Ziziphus mauritiana Lam.) and Mining Differentially Expressed Genes Related to the Tolerance through Transcriptome Analysis
Abstract
:1. Introduction
2. Results
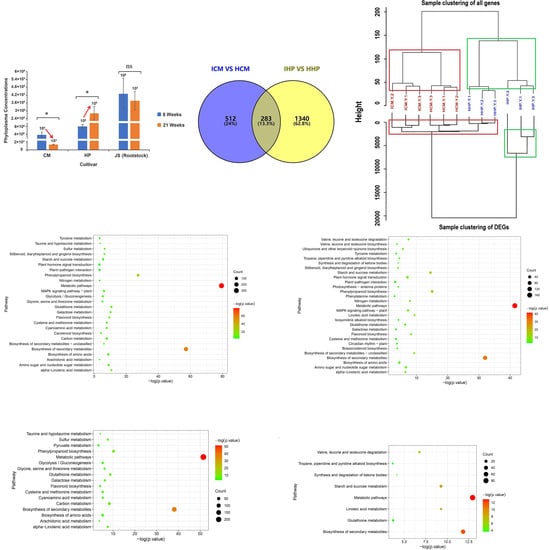
2.1. Indian Jujube ‘Cuimi’ Is High-Tolerant to JWB
2.2. Screening for Differentially Expressed Genes in ‘Cuimi’ and ‘Huping’
2.3. Gene Ontology (GO) Enrichment Analysis of DEGs
2.4. Identification of Genes Associated with ‘Cuimi’ High Tolerance
3. Discussion
3.1. The High Tolerance to JWB of ‘Cuimi’
3.2. Response of JWB Susceptible Jujube Cultivar to Phytoplasma Infection
3.3. Response of JWB Tolerant Jujube Cultivar to Phytoplasma Infection
4. Materials and Methods
4.1. Plant Materials
4.2. Detection of JWB Phytoplasma Using Nested PCR
4.3. Quantitative PCR (qPCR)
4.4. RNA-Seq Library Construction and Sequencing
4.5. Data Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jung, H.Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Wei, W.; Oshima, K.; Miyata, S.I.; Ugaki, M.; Hibi, T.; Namba, S. ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’-broom disease. Int. J. Syst. Evol. Microbiol. 2003, 53, 1037–1041. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhao, Z.H. Germplasm Resources and Production of Jujube in China. Acta Hortic. 2009, 840, 25–32. [Google Scholar] [CrossRef]
- Tian, G.Z.; Zhang, Z.S.; Li, Z.Q.; Guo, J.H. Dynamic of Jujube Witches’ Broom Disease and Factors of Great Influence at Ecologically Different Regions in China. Sci. Silvae Sin. 2002, 38, 83–91. [Google Scholar]
- Bertaccini, A.; Duduk, B.; Paltrinieri, S.; Contaldo, N. Phytoplasmas and Phytoplasma Diseases: A Severe Threat to Agriculture. Am. J. Plant. Sci. 2014, 5, 1763–1788. [Google Scholar] [CrossRef]
- Bertaccini, A. Containment of Phytoplasma-Associated Plant Diseases by Antibiotics and Other Antimicrobial Molecules. Antibiotics 2021, 10, 1398. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, Z.C.; Liu, M.J. The field resistant character of Ziziphus jujuba Mill ‘Xingguang’ with high resistence to jujube witches’ broom. Acta Phytophylacica Sin. 2010, 37, 89–90. [Google Scholar] [CrossRef]
- Wang, L.; Liu, S.; Gao, M.; Wang, L.; Wang, L.; Wang, Y.; Dai, L.; Zhao, J.; Liu, M.; Liu, Z. The Crosstalk of the Salicylic Acid and Jasmonic Acid Signaling Pathways Contributed to Different Resistance to Phytoplasma Infection Between the Two Genotypes in Chinese Jujube. Front. Microbiol. 2022, 13, 800762. [Google Scholar] [CrossRef]
- Ramadugu, C.; Keremane, M.L.; Halbert, S.E.; Duan, Y.P.; Roose, M.L.; Stover, E.; Lee, R.F. Long-Term Field Evaluation Reveals Huanglongbing Resistance in Citrus Relatives. Plant Dis. 2016, 100, 1858–1869. [Google Scholar] [CrossRef]
- Sun, Z.J. Paliurus hemsleyanus Rehder used as jujube rootstock for 30 years. Econ. For. Res. 1992, S1, 159–160. [Google Scholar] [CrossRef]
- Sobhani, Z.; Nikoofal-Sahlabadi, S.; Amiri, M.S.; Ramezani, M.; Emami, S.A.; Sahebkar, A. Therapeutic Effects of Ziziphus jujuba Mill. Fruit in Traditional and Modern Medicine: A Review. Med. Chem. 2020, 16, 1069–1088. [Google Scholar] [CrossRef]
- Tahergorabi, Z.; Abedini, M.R.; Mitra, M.; Fard, M.H.; Beydokhti, H. “Ziziphus jujuba”: A red fruit with promising anticancer activities. Pharmacogn. Rev. 2015, 9, 99–106. [Google Scholar] [CrossRef]
- Chiou, C.-Y.; Shih, H.-C.; Tsai, C.-C.; Jin, X.-L.; Ko, Y.-Z.; Mantiquilla, J.A.; Weng, I.-S.; Chiang, Y.-C. The genetic relationships of Indian jujube (Ziziphus mauritiana Lam.) cultivars using SSR markers. Heliyon 2020, 6, e05078. [Google Scholar] [CrossRef]
- Jamadar, M.M.; Balikai, R.; Sataraddi, A.R. Status of Diseases on Ber (Ziziphus mauritiana lamarck) in India and their Management Options. Acta Hortic. 2009, 840, 383–390. [Google Scholar] [CrossRef]
- Kumar, J.; Ramlal, A.; Kumar, K.; Rani, A.; Mishra, V. Signaling Pathways and Downstream Effectors of Host Innate Immunity in Plants. Int. J. Mol. Sci. 2021, 22, 9022. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Gao, P.; Duan, T.; Christensen, M.; Nan, Z.; Liu, Q.; Meng, F.; Huang, J. The occurrence of rust disease, and biochemical and physiological responses on Apocynum venetum plants grown at four soil water contents, following inoculation with Melampsora apocyni. Eur. J. Plant Pathol. 2018, 150, 549–563. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Xiao, J.; Zhao, J.; Liu, M. Identification of genes associated with phytoplasma resistance through suppressive subtraction hybridization in Chinese jujube. Physiol. Mol. Plant Pathol. 2014, 86, 43–48. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, J.; Liu, M. Photosynthetic responses to phytoplasma infection in Chinese jujube. Plant Physiol. Biochem. 2016, 105, 12–20. [Google Scholar] [CrossRef]
- Ma, F.; Huang, J.; Yang, J.; Zhou, J.; Sun, Q.; Sun, J. Identification, expression and miRNA targeting of auxin Response Factor Genes Relat. to phyllody in the witches’ broom disease of jujube. Gene 2020, 746, 144656. [Google Scholar] [CrossRef]
- Wang, H.; Ye, X.; Li, J.; Tan, B.; Chen, P.; Cheng, J.; Wang, W.; Zheng, X.; Feng, J. Transcriptome profiling analysis revealed co-regulation of multiple pathways in jujube during infection by ‘Candidatus Phytoplasma ziziphi’. Gene 2018, 665, 82–95. [Google Scholar] [CrossRef]
- Xue, C.; Liu, Z.; Dai, L.; Bu, J.; Liu, M.; Zhao, Z.; Jiang, Z.; Gao, W.; Zhao, J. Changing Host Photosynthetic, Carbohydrate, and Energy Metabolisms Play Important Roles in Phytoplasma Infection. Phytopathology 2018, 108, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Z.; Zhang, C.; Li, H.; Li, X. Transcriptomic Analysis on Pathogenesis of Witches’ Broom Disease in Ziziphus jujuba. Acta Hortic. Sin. 2017, 44, 1287–1298. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, F.; Yao, Y.; Deng, M.; Chen, M.; Zhang, S.; Li, Y.; Yang, J.; Zhang, N.; Huang, J.; et al. Jujube witches’ broom phytoplasma effectors SJP1 and SJP2 induce lateral bud outgrowth by repressing the ZjBRC1-controlled auxin efflux channel. Plant Cell Environ. 2021, 44, 3257–3272. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Patui, S.; Bertolini, A.; Clincon, L.; Ermacora, P.; Braidot, E.; Vianello, A.; Zancani, M. Involvement of plasma membrane peroxidases and oxylipin pathway in the recovery from phytoplasma disease in apple (Malus domestica). Physiol. Plant. 2013, 148, 200–213. [Google Scholar] [CrossRef]
- Buta, J.G.; Spaulding, D.W. Changes in indole-3-acetic acid and abscisic acid levels during tomato (Lycopersicon esculentum Mill.) fruit development and ripening. J. Plant Growth Regul. 1994, 13, 163–166. [Google Scholar] [CrossRef]
- Chen, P.; Chen, L.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Yang, Q.; Zhang, Y.; Li, J.; et al. Phytoplasma effector Zaofeng6 induces shoot proliferation by decreasing the expression of ZjTCP7 in Ziziphus jujuba. Hortic. Res. 2022, 9, uhab032. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant-Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef]
- Lee, I.M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef]
- Ye, X.; Wang, H.; Chen, P.; Fu, B.; Zhang, M.; Li, J.; Zheng, X.; Tan, B.; Feng, J. Combination of iTRAQ proteomics and RNA-seq transcriptomics reveals multiple levels of regulation in phytoplasma-infected Ziziphus jujuba Mill. Hortic. Res. 2017, 4, 17080. [Google Scholar] [CrossRef]
- Lepka, P.; Stitt, M.; Moll, E.; Seemüller, E. Effect of phytoplasmal infection on concentration and translocation of carbohydrates and amino acids in periwinkle and tobacco. Physiol. Mol. Plant Pathol. 1999, 55, 59–68. [Google Scholar] [CrossRef]
- Guthrie, J.N.; Walsh, K.B.; Scott, P.T.; Rasmussen, T.S. The phytopathology of Australian papaya dieback: A proposed role for the phytoplasma. Physiol. Mol. Plant Pathol. 2001, 58, 23–30. [Google Scholar] [CrossRef]
- Maust, B.E.; Espadas, F.; Talavera, C.; Aguilar, M.; Santamaría, J.M.; Oropeza, C. Changes in carbohydrate metabolism in coconut palms infected with the lethal yellowing phytoplasma. Phytopathology 2003, 93, 976–981. [Google Scholar] [CrossRef]
- Junqueira, A.; Bedendo, I.; Pascholati, S. Biochemical changes in corn plants infected by the maize bushy stunt phytoplasma. Physiol. Mol. Plant Pathol. 2004, 65, 181–185. [Google Scholar] [CrossRef]
- Oshima, K.; Kakizawa, S.; Nishigawa, H.; Jung, H.Y.; Wei, W.; Suzuki, S.; Arashida, R.; Nakata, D.; Miyata, S.; Ugaki, M.; et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 2004, 36, 27–29. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.-J.; Reddy, M.S.S.; Wang, L. The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Wang, H.; Ye, X.; Li, J.; Tan, B.; Chen, P.; Jiang, Y.; Cheng, J.; Wang, W.; Zheng, X.; Feng, J. Combination of iTRAQ proteomics and RNA-seq transcriptomics reveals jasmonate-related-metabolisms central regulation during the process of Jujube witches’ broom recovery by tetracycline treatment. Sci. Hortic. 2019, 243, 197–206. [Google Scholar] [CrossRef]
- Xue, C.; Li, H.; Liu, Z.; Wang, L.; Zhao, Y.; Wei, X.; Fang, H.; Liu, M.; Zhao, J. Genome-wide analysis of the WRKY gene family and their positive responses to phytoplasma invasion in Chinese jujube. BMC Genom. 2019, 20, 464. [Google Scholar] [CrossRef]
- Tena, G.; Boudsocq, M.; Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011, 14, 519–529. [Google Scholar] [CrossRef]
- Gao, Y.-L.; Wang, B.-W.; Xu, Z.-L.; Li, M.-Y.; Song, Z.-B.; Li, W.-Z.; Li, Y.-P. Tobacco serine/threonine protein kinase gene NrSTK enhances black shank resistance. Genet. Mol. Res. 2015, 14, 16415–16424. [Google Scholar] [CrossRef]
- Lin, Z.J.; Liebrand, T.W.; Yadeta, K.A.; Coaker, G. PBL13 Is a Serine/Threonine Protein Kinase That Negatively Regulates Arabidopsis Immune Responses. Plant Physiol. 2015, 169, 2950–2962. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Tanaka, K.; Poovaiah, B.W. Calcium/Calmodulin-Mediated Defense Signaling: What Is Looming on the Horizon for AtSR1/CAMTA3-Mediated Signaling in Plant Immunity. Front. Plant Sci. 2021, 12, 795353. [Google Scholar] [CrossRef] [PubMed]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Chiasson, D.; Ekengren, S.K.; Martin, G.B.; Dobney, S.L.; Snedden, W.A. Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 2005, 58, 887–897. [Google Scholar] [CrossRef]
- Quentin, M.; Allasia, V.; Pegard, A.; Allais, F.; Ducrot, P.H.; Favery, B.; Levis, C.; Martinet, S.; Masur, C.; Ponchet, M.; et al. Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog. 2009, 5, e1000264. [Google Scholar] [CrossRef]
- Bart, R.S.; Chern, M.; Vega-Sánchez, M.E.; Canlas, P.; Ronald, P.C. Rice Snl6, a cinnamoyl-CoA reductase-like gene family member, is required for NH1-mediated immunity to Xanthomonas oryzae pv. oryzae. PLoS Genet. 2010, 16, e1001123. [Google Scholar] [CrossRef]
- Duduk, B.; Paltrinieri, S.; Lee, I.M.; Bertaccini, A. Nested PCR and RFLP analysis based on the 16S rRNA gene. In Phytoplasma. Methods in Molecular Biology; Dickinson, M., Hodgetts, J., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 159–171. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, H.; Lin, C.; Liu, X.; Song, C.; Feng, S.; Yu, S.; Lu, X.; Tian, G. A real-time (SYBR Green I) PCR assay for detection and quantification of jujube witches’-broom phytoplasma in the grafted jujube cultivar scions with different resistance. Acta Phytopathol. Sin. 2015, 45, 520–529. [Google Scholar]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Wang, C.; Kong, D.; Cao, M.; Zhang, Q.; Tahir, M.; Yang, Y.; Yang, S.; Bo, W.; Pang, X. Identification of High Tolerance to Jujube Witches’ Broom in Indian Jujube (Ziziphus mauritiana Lam.) and Mining Differentially Expressed Genes Related to the Tolerance through Transcriptome Analysis. Plants 2023, 12, 2082. https://doi.org/10.3390/plants12112082
Xu Y, Wang C, Kong D, Cao M, Zhang Q, Tahir M, Yang Y, Yang S, Bo W, Pang X. Identification of High Tolerance to Jujube Witches’ Broom in Indian Jujube (Ziziphus mauritiana Lam.) and Mining Differentially Expressed Genes Related to the Tolerance through Transcriptome Analysis. Plants. 2023; 12(11):2082. https://doi.org/10.3390/plants12112082
Chicago/Turabian StyleXu, Yaru, Chao Wang, Decang Kong, Ming Cao, Qiong Zhang, Muhammad Tahir, Ying Yang, Shuang Yang, Wenhao Bo, and Xiaoming Pang. 2023. "Identification of High Tolerance to Jujube Witches’ Broom in Indian Jujube (Ziziphus mauritiana Lam.) and Mining Differentially Expressed Genes Related to the Tolerance through Transcriptome Analysis" Plants 12, no. 11: 2082. https://doi.org/10.3390/plants12112082