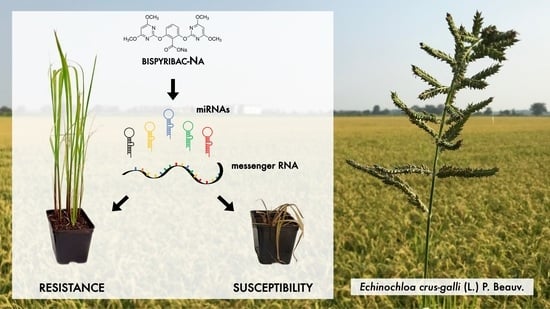
Involvement of miRNAs in Metabolic Herbicide Resistance to Bispyribac-Sodium in Echinochloa crus-galli (L.) P. Beauv.
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Herbicide Treatment
4.2. RNA Extraction
4.3. Candidate NTSR Gene Selection and Candidate miRNA Prediction
4.4. cDNA Synthesis and qRT-PCR Analysis
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef]
- Petit, B.; Duhieu, K.; Boucansaud, C.; Délye, C. Complex genetic control of non-target site based resistance to herbicides inhibiting acetyl-coenzyme A carboxylase and acetolactate synthase in Alopecurus myosuroides Huds. Plant Sci. 2010, 178, 501–509. [Google Scholar] [CrossRef]
- Iwakami, S.; Uchino, A.; Kataoka, Y.; Shibaike, H.; Watanabe, H.; Inamura, T. Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest Manag. Sci. 2013, 70, 549–558. [Google Scholar] [CrossRef]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 Are Associated with Resistance to Two Acetolactate Synthase Inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [Green Version]
- Jugulam, M.; Shyam, C. Non-Target-Site Resistance to Herbicides: Recent Developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, A.; Rodriguez-Carres, M.; Sasidharan, R.; Koski, L.; Peterson, D.; Nandula, V.; Ray, J.D.; Bond, J.; Shaw, D. Multiple Herbicide–Resistant Junglerice (Echinochloa colona): Identification of Genes Potentially Involved in Resistance through Differential Gene Expression Analysis. Weed Sci. 2018, 66, 347–354. [Google Scholar] [CrossRef]
- Yuan, J.S.; Tranel, P.J.; Stewart, C.N., Jr. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L.; et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017, 8, 1031. [Google Scholar] [CrossRef] [Green Version]
- Panozzo, S.; Scarabel, L.; Rosan, V.; Sattin, M. A new ala-122-Asn amino acid change confers decreased fitness to ALS-resistant Echinochloa crus-galli. Front. Plant Sci. 2017, 8, 2042. [Google Scholar] [CrossRef] [Green Version]
- Panozzo, S.; Mascanzoni, E.; Scarabel, L.; Milani, A.; Dalazen, G.; Merotto, A.J.; Tranel, P.J.; Sattin, M. Target-Site Mutations and Expression of ALS Gene Copies Vary According to Echinochloa Species. Genes 2021, 12, 1841. [Google Scholar] [CrossRef]
- Panozzo, S.; Scarabel, L.; Tranel, P.J.; Sattin, M. Target-site resistance to ALS inhibitors in the polyploid species Echinochloa crus-galli. Pestic. Biochem. Physiol. 2013, 105, 93–101. [Google Scholar] [CrossRef]
- Mascanzoni, E.; Perego, A.; Marchi, N.; Scarabel, L.; Panozzo, S.; Ferrero, A.; Acutis, M.; Sattin, M. Epidemiology and agronomic predictors of herbicide resistance in rice at a large scale. Agron. Sustain. Dev. 2018, 38, 68. [Google Scholar] [CrossRef] [Green Version]
- McElroy, J. Vavilovian Mimicry: Nikolai Vavilov and His Little-Known Impact on Weed Science. Weed Sci. 2014, 62, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Ye, C.Y.; Tang, W.; Wu, D.; Jia, L.; Qiu, J.; Chen, M.; Mao, L.; Lin, F.; Xu, H.; Yu, X.; et al. Genomic evidence of human selection on Vavilovian mimicry. Nat. Ecol. Evol. 2019, 3, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Guo, Q.; Wang, J.; Shi, L.; Yang, X.; Zhou, Y.; Yu, Q.; Bai, L. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli. J. Hazard. Mater. 2022, 428, 128225. [Google Scholar] [CrossRef] [PubMed]
- Vidotto, F.; Tesio, F.; Tabacchi, M.; Ferrero, A. Herbicide sensitivity of Echinochloa spp. accessions in Italian rice fields. Crop Protect. 2007, 26, 285–293. [Google Scholar] [CrossRef]
- Brusoni, M. Studio sulla variabilità intraspecifica di Echinochloa crus-galli (L.) P. Beauv. Atti. Ist. Bot. Lab. Crittogam. 1991, 10, 39–88. [Google Scholar]
- Cusaro, C.M.; Grazioli, C.; Zambuto, F.; Capelli, E.; Brusoni, M. An Improved Method for Assessing Simple Sequence Repeat (SSR) Variation in Echinochloa crus-galli (L.) P. Beauv (Barnyardgrass). Diversity 2022, 14, 3. [Google Scholar] [CrossRef]
- Norris, R.F. Morphological and Phenological Variation in Barnyardgrass (Echinochloa crus-galli) in California. Weed Sci. 1996, 44, 804–814. [Google Scholar] [CrossRef]
- Rutledge, J.; Talbert, R.; Sneller, C. RAPD analysis of genetic variation among propanil-resistant and -susceptible Echinochloa crus-galli populations in Arkansas. Weed Sci. 2000, 48, 669–674. [Google Scholar] [CrossRef]
- Tasrif, A.; Juraimi, A.S.; Kadir, J.; Sastroutomo, S.S.; Napis, S. Genetic diversity of Echinochloa crus-galli var. crus-galli (L.) Beauv (Barnyardgrass: Poaceae) ecotypes in Malaysia and Indonesia as revealed by RAPD markers. Asian J. Plant Sci. 2004, 3, 231–238. Asian J. Plant Sci. 2004, 3, 231–238. [Google Scholar] [CrossRef]
- Altop, E.K.; Mennan, H. Genetic and morphologic diversity of Echinochloa crus-galli populations from different origins. Phytoparasitica 2010, 39, 93–102. [Google Scholar] [CrossRef]
- Lee, J.; Park, K.W.; Lee, I.Y.; Kim, C.S.; Kown, O.D.; Park, T.S. Simple sequence repeat analysis of genetic diversity among Acetyl-CoA carboxylase inhibitor-resistant and inhibitor-susceptible Echinochloa crus-galli and E. oryzicola populations in Korea. Weed Res. 2015, 55, 90–100. [Google Scholar] [CrossRef]
- Le, D.; Nguyen, C.M.; Mann, R.K.; Yerkes, C.N.; Kumar, B.V.N. Genetic diversity and herbicide resistance of 15 Echinochloa crus-galli populations to quinclorac in Mekong Delta of Vietnam and Arkansas of United States. J. Plant Biotechnol. 2017, 44, 472–477. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Zhang, W.; Fang, J.; Dong, L. Restriction site-associated DNA sequencing allows for the rapid identification of simple sequence repeat markers in Echinochloa crus-galli. Weed Biol. Manag. 2017, 17, 68–76. [Google Scholar] [CrossRef]
- Nozawa, H.; Takahashi, M.; Nakai, H.; Sato, Y. Difference in SSR Variations Between Japanese Barnyard Millet (Echinochloa esculenta) and its Wild Relative E. crus-galli. Breed. Sci. 2006, 56, 335–340. [Google Scholar] [CrossRef] [Green Version]
- Oerke, E.C.; Dehne, H.W. Safeguarding production-losses in major crops and the role of crop protection. Crop Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- Riar, D.S.; Norsworthy, J.K.; Srivastava, V.; Nandula, V.; Bond, J.A.; Scott, R.C. Physiological and molecular basis of acetolactate synthase-inhibiting herbicide resistance in barnyardgrass (Echinochloa crus-galli). J. Agric. Food Chem. 2013, 61, 278–289. [Google Scholar] [CrossRef]
- Matzenbacher, F.O.; Bortoly, E.D.; Kalsing, A.; Merotto, A. Distribution and analysis of the mechanisms of resistance of barnyardgrass (Echinochloa crus-galli) to imidazolinone and quinclorac herbicides. J. Agric. Sci. 2015, 153, 1044–1058. [Google Scholar] [CrossRef]
- Dalazen, G.; Markus, C.; Merotto, A., Jr. Differential Expression of Genes Associated with Degradation Enhancement of Imzethapyr in Barnyardgrass (Echinochloa crus-galli). J. Agric. Sci. 2018, 10, 389–401. [Google Scholar] [CrossRef]
- Fang, J.; Zhang, Y.; Liu, T.; Yan, B.; Li, J.; Dong, L. Target-Site and Metabolic Resistance Mechanisms to Penoxsulam in Barnyardgrass (Echinochloa crus-galli (L.) P. Beauv). J. Agric. Food Chem. 2019, 67, 8085–8095. [Google Scholar] [CrossRef]
- Markus, C.; Pecinka, A.; Karan, R.; Barney, J.N.; Merotto, A. Epigenetic regulation-contribution to herbicide resistance in weeds? Pest Manag. Sci. 2018, 74, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Margaritopoulou, T.; Tani, E.; Chachalis, D.; Travlos, I. Involvement of Epigenetic Mechanisms in Herbicide Resistance: The Case of Conyza canadensis. Agriculture 2018, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Goh, S.S.; Yu, Q.; Han, H.; Vila-Aiuba, M.M.; Busia, R.; Powles, S.B. Non-target-site glyphosate resistance in Echinochloa colona from Western Australia. Crop Prot. 2018, 112, 257–263. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Sun, G. MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 2012, 80, 17–36. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Leung, A.K.; Sharp, P.A. MicroRNA functions in stress responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Xue, L.; An, L. Functional diversity of miRNA in plants. Plant Sci. 2007, 172, 423–432. [Google Scholar] [CrossRef]
- Zhao, W.; Xiao, W.; Sun, J.; Chen, M.; Ma, M.; Cao, Y.; Cen, W.; Li, R.; Luo, J. An Integration of MicroRNA and Transcriptome Sequencing Analysis Reveal Regulatory Roles of miRNAs in Re-sponse to Chilling Stress in Wild Rice. Plants 2022, 11, 977. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, 31–36. [Google Scholar] [CrossRef]
- Mathur, M.; Nair, A.; Kadoo, N. Plant-pathogen interactions: MicroRNA-mediated trans-kingdom gene regulation in fungi and their host plants. Genomics 2020, 112, 3021–3035. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Waterhouse, P.M. Plant and animal microRNAs: Similarities and differences. Funct. Integr. Genom. 2005, 5, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Arteaga-Vázquez, M.; Caballero-Pérez, J.; Vielle-Calzada, J.P. A family of microRNAs present in plants and animals. Plant Cell 2006, 18, 3355–3369. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zhang, B. MicroRNAs in Control of Plant Development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Llave, C. MicroRNAs: More than a role in plant development? Mol. Plant Pathol. 2004, 5, 361–366. [Google Scholar] [CrossRef]
- Sun, X.; Lin, L.; Sui, N. Regulation mechanism of microRNA in plant response to abiotic stress and breeding. Mol. Biol. Rep. 2019, 46, 1447–1457. [Google Scholar] [CrossRef]
- Jatan, R.; Lata, C. Role of MicroRNAs in Abiotic and Biotic Stress Resistance in Plants. Proc. Indian Natl. Sci. Acad 2019, 85, 553–567. [Google Scholar]
- Chauhan, S.; Yogindran, S.; Rajam, M.V. Role of miRNAs in biotic stress reactions in plants. Indian J. Plant Physiol. 2017, 22, 514–529. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Iwakami, S. Cytochrome P450-mediated herbicide metabolism in plants: Current understanding and prospects. Pest Manag. Sci. 2021, 77, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Wang, Z.; Cai, J.; Gao, H.; Zhao, H.; Dong, L. High-throughput sequencing reveals differential regulation of miRNAs in fenoxaprop-P-ethyl-resistant Beckmannia syzigachne. Sci. Rep. 2016, 6, 28725. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Wang, Y.; Yao, Y.; Bai, Y.; Wu, K.; Qiao, Y. Identification microRNAs and target genes in Tibetan hulless barley to BLS infection. Agron. J. 2021, 113, 2273–2292. [Google Scholar] [CrossRef]
- Li, G.; Wu, S.G.; Yu, R.X.; Cang, T.; Chen, L.P.; Zhao, X.P.; Cai, L.M.; Wu, C.X. Identification and expression pattern of a gluta- thione S-transferase in Echinochloa crus-galli. Weed Res. 2013, 53, 314–321. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhuang, Z.; Zhao, P.X. Computational analysis of miRNA targets in plants: Current status and challenges. Brief. Bioinform. 2011, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Mehra, S.; Chatterjee, S.; Purty, R.S. In silico identification and validation of miRNA and their DIR specific targets in Oryza sativa Indica under abiotic stress. Non-Coding RNA Res. 2020, 5, 167–177. [Google Scholar] [CrossRef]
- Fang, C.; Li, Y.; Li, C.; Li, B.; Ren, Y.; Zheng, H.; Zeng, X.; Shen, L.; Lin, W. Identification and comparative analysis of microRNAs in barnyardgrass (Echinochloa crus-galli) in response to rice allelopathy. Plant Cell Environ. 2015, 38, 1368–1381. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, S.; Kong, X.; Li, Y.; Zhao, G.; He, W.; Appels, R.; Pfeifer, M.; Tao, Y.; Zhang, X.; et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 2013, 496, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Glazińska, P.; Kulasek, M.; Glinkowski, W.; Wojciechowski, W.; Kosiński, J. Integrated Analysis of Small RNA, Transcriptome and Degradome Sequencing Provides New Insights into Floral Development and Abscission in Yellow Lupine (Lupinus luteus L.). Int. J. Mol. Sci. 2019, 20, 5122. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-J.; Guo, H.-S. Cleavage of Indole-3-Acetic Acid Inducible28 mRNA by microRNA847 upregulates auxin signaling to modulate cell proliferation and lateral organ growth in Arabidopsis. Plant Cell 2015, 27, 574–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, W.; Wang, Z.; Du, M.; Liu, Y.; Liu, J.Y. Genome-wide analysis of small RNAs reveals eight fiber elongation-related and 257 novel microRNAs in elongating cotton fiber cells. BMC Genom. 2013, 14, 629. [Google Scholar] [CrossRef]
- Shi, Y.; Xia, H.; Cheng, X.; Zhang, L. Genome-wide miRNA analysis and integrated net-work for flavonoid biosynthesis in Osmanthus fragrans. BMC Genom. 2021, 22, 141. [Google Scholar] [CrossRef]
- Li, A.L.; Wen, Z.; Yang, K.; Wen, X.P. Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2019, 20, 4795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthewman, C.A.; Kawashima, C.G.; Húska, D.; Csorba, T.; Dalmay, T.; Kopriva, S. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 2012, 586, 3242–3248. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radwan, O.; Liu, Y.; Clough, S.J. Transcriptional analysis of soybean root response to Fusarium virguliforme, the causal agent of sudden death syndrome. Mol. Plant-Microbe Interact. MPMI 2011, 24, 958–972. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.Q.; Yan, L.F.; Wang, T. Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 2011, 12, R53. [Google Scholar] [CrossRef] [Green Version]
- Salanoubat, M.; Lemcke, K.; Rieger, M.; Ansorge, W.; Unseld, M.; Fartmann, B.; Valle, G.; Blöcker, H.; Perez-Alonso, M.; Ober-maier, B.; et al. Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 2000, 408, 820–823. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; De Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; Rodrigues, F.A.; Marcolino, J.; Barbosa, J.F.; Stolf-Moreira, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaMonte, G.; Philip, N.; Reardon, J.; Lacsina, J.R.; Majoros, W.; Chapman, L.; Thornburg, C.D.; Telen, M.J.; Ohler, U.; Nicchitta, C.V.; et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012, 12, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Luo, R.; Hu, C.; Cao, J.; Jin, Y. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology 2013, 140, 1751–1761. [Google Scholar] [CrossRef]
- Shahid, S.; Kim, G.; Johnson, N.R.; Wafula, E.; Wang, F.; Coruh, C.; Bernal-Galeano, V.; Phifer, T.; de Pamphilis, C.W.; Westwood, J.H.; et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553, 82–85. [Google Scholar] [CrossRef]
- Zhang, L.L.; Jing, X.D.; Chen, W.; Wang, Y.; Lin, J.H.; Zheng, L.; Dong, Y.H.; Zhou, L.; Li, F.F.; Yang, F.Y.; et al. Host plant-derived miRNAs potentially modulate the development of a cosmopolitan insect pest, Plutella xylostella. Biomolecules 2019, 9, 602. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Jing, X.; Chen, W.; Bai, J.; Vasseur, L.; He, W.; You, M. Selection of reference genes for expression analysis of plant-derived microRNAs in Plutella xylostella using qRT-PCR and ddPCR. PLoS ONE 2019, 14, e0220475. [Google Scholar] [CrossRef] [Green Version]
- EPPO. Efficacy evaluation of herbicides: Weeds in water-seeded rice. Bulletin 2011, 41, 282–285. Available online: https://pp1.eppo.int/standards/PP1-062-3 (accessed on 16 April 2022).
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 9 September 2022).



| miRNAs | Target Genes | ||
|---|---|---|---|
| Name | a.n. (miRbase) | Name | a.n. (NCBI) |
| ata-miR166c-5p | MIMAT0037248 | CYP72A122 | AB734013.1 |
| ath-miR396b-5p | MIMAT0000945 | CYP81A22 | AB872310.1 |
| osa-miR395f | MIMAT0000974 | CYP81A68 | OK483200.1 |
| ath-miR847 | MIMAT0004278 | CYP71AK2 | AB733990.1 |
| gra-miR7486c | MIMAT0034235 | CYP72A254 | AB755796.1 |
| gma-miR396f | MIMAT0021069 | GSTF1 | HF548530.1 |
| osa-miR5538 | MIMAT0022174 | EcGST1 | JX518596 |
| gra-miR8759 | MIMAT0034189 | eIF4B1 | AB720070.1 |
| Gene ID | NCBI a.n. | Primer Sequence (5′-3′) | Reference |
|---|---|---|---|
| CYP71AK2 | AB733990.1 | F: acgtgtgggacaagttcctg | Iwakami et al., 2013 [3] |
| R: ggctttgatgcgatcgtctg | |||
| CYP72A254 | AB755796.1 | F: ttacgaggtactccggctgt | Iwakami et al., 2013 [3] |
| R: gtcagggtcgtggtgaatgt | |||
| CYP72A122 | AB734013.1 | F: agttcaagccggagaggttc | Iwakami et al., 2013 [3] |
| R: catcttggcttcaagcagcg | |||
| CYP81A68 | OK483200.1 | F: gactattcaacccgggcgat | Pan et al., 2022 [15] |
| R: caagttctgcacggcaagag | |||
| CYP81A22 | AB872310.1 | F: cggcgcgctggtccagtt | Iwakami et al., 2014 [4] |
| R: tgacatgagcagttccatcg | |||
| EcGST1 | JX518596 | F: gccgaggaggacctgaagaac | Li et al., 2013 [54] |
| R: gtgactcacagataggcttaccgt | |||
| GSTF1 | HF548530.1 | F: tgcctcttcaaccccatgat | Dalazen et al., 2018 [30] |
| R: aggtactcgtgctgggagag | |||
| eIF4B1 | AB720070.1 | F: cgagcagcttacaagggact | Dalazen et al., 2018 [30] |
| R: gtggttccataccaccacga | |||
| b-actin | HQ395760.1 | F: gtgctgttccagccatcgttcat | Li et al., 2013 [54] |
| R: ctccttgctcatacggtcagcaata |
| Name | miRbase a.n. | miRNA Sequence (5′-3′) | Reference |
|---|---|---|---|
| ata-miR166c-5p | MIMAT0037248 | ggaacguuggcuggcucgagg | Jia et al., 2013 [59] |
| ath-miR396b | MIMAT0000945 | uuccacagcuuucuugaacuu | John-Rohades et al., 2004 [66] |
| ath-miR847 | MIMAT0004278 | ucacuccucuucuucuugaug | Rajagopalan et al., 2006 [67] |
| gma-miR396f | MIMAT0021069 | agcuuucuugaacuucuuaugccua | Radwan et al., 2011 [68] |
| gra-miR7486c | MIMAT0034235 | uuuguccacgugaacagaaaacgc | Xue et al., 2013 [62] |
| gra-miR8759 | MIMAT0034189 | ugguggaaguauugugcccgg | Xue et al., 2013 [62] |
| osa-miR395f | MIMAT0000974 | gugaauuguuugggggaacuc | John-Rohades et al., 2004 [66] |
| osa-miR5538 | MIMAT0022174 | acugaacucaaucacuugcugc | Wei et al., 2011 [69] |
| U6 snRNA | NR141593.1 | cttcggggacatccgataaaattg | Salanoubat et al., 2000 [70] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cusaro, C.M.; Grazioli, C.; Capelli, E.; Picco, A.M.; Guarise, M.; Gozio, E.; Zarpellon, P.; Brusoni, M. Involvement of miRNAs in Metabolic Herbicide Resistance to Bispyribac-Sodium in Echinochloa crus-galli (L.) P. Beauv. Plants 2022, 11, 3359. https://doi.org/10.3390/plants11233359
Cusaro CM, Grazioli C, Capelli E, Picco AM, Guarise M, Gozio E, Zarpellon P, Brusoni M. Involvement of miRNAs in Metabolic Herbicide Resistance to Bispyribac-Sodium in Echinochloa crus-galli (L.) P. Beauv. Plants. 2022; 11(23):3359. https://doi.org/10.3390/plants11233359
Chicago/Turabian StyleCusaro, Carlo Maria, Carolina Grazioli, Enrica Capelli, Anna Maria Picco, Marta Guarise, Enrico Gozio, Pietro Zarpellon, and Maura Brusoni. 2022. "Involvement of miRNAs in Metabolic Herbicide Resistance to Bispyribac-Sodium in Echinochloa crus-galli (L.) P. Beauv." Plants 11, no. 23: 3359. https://doi.org/10.3390/plants11233359