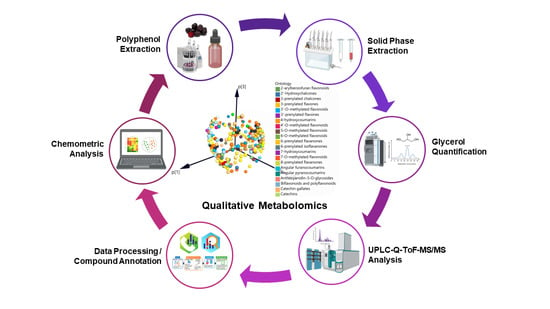
Qualitative Analysis of Polyphenols in Glycerol Plant Extracts Using Untargeted Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Material and Extraction
2.3. Glycerol Removal Using Solid Phase Extraction (SPE)
2.4. Glycerol Quantification
2.5. HPLC-RID Method Validation
2.6. UPLC-Q-ToF MS/MS Analysis
2.7. Processing of LC-MS/MS Data
2.8. Statistical Analysis
3. Results
3.1. HPLC-RID Glycerol Quantification
3.2. Untargeted Metabolic Profiling of QGP Extracts Using UPLC-Q-ToF-MS-MS
3.3. Characterisation and Comparison of Abundant Polyphenols
3.4. Qualitative Analysis of Polyphenols in Queen Garnet Plum
4. Discussion
4.1. Glycerol Removal for UPLC-Q-ToF-MS/MS Analysis
4.2. Untargeted Metabolomics of Queen Garnet Plum Polyphenols
4.3. Significance of the Results and Future Applications of Glycerol Plant Extracts
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef] [PubMed]
- McLafferty, F.W. Tandem mass spectrometry. Science 1981, 214, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Boesl, U. Time-of-flight mass spectrometry: Introduction to the basics. Mass Spectrom. Rev. 2017, 36, 86–109. [Google Scholar] [CrossRef]
- Chernushevich, I.V.; Loboda, A.V.; Thomson, B.A. An introduction to quadrupole–time-of-flight mass spectrometry. J. Mass Spectrom. 2001, 36, 849–865. [Google Scholar] [CrossRef]
- Pan, X.; Welti, R.; Wang, X. Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography–electrospray tandem mass spectrometry. Phytochemistry 2008, 69, 1773–1781. [Google Scholar] [CrossRef]
- Fang, C.; Fernie, A.R.; Luo, J. Exploring the diversity of plant metabolism. Trends Plant Sci. 2019, 24, 83–98. [Google Scholar] [CrossRef]
- Gertsman, I.; Barshop, B.A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metab. Dis. 2018, 41, 355–366. [Google Scholar] [CrossRef]
- Misra, B.B. New software tools, databases, and resources in metabolomics: Updates from 2020. Metabolomics 2021, 17, 49. [Google Scholar] [CrossRef]
- Alonso, A.; Marsal, S.; Julià, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef] [PubMed]
- Fraisier-Vannier, O.l.; Chervin, J.; Cabanac, G.; Puech, V.; Fournier, S.; Durand, V.; Amiel, A.L.; André, O.; Benamar, O.A.; Dumas, B. MS-CleanR: A feature-filtering workflow for untargeted LC–MS based metabolomics. Anal. Chem. 2020, 92, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- Suber, P. Open Access; The MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Pavlić, B.; Mrkonjić, Ž.; Teslić, N.; Kljakić, A.C.; Pojić, M.; Mandić, A.; Stupar, A.; Santos, F.; Duarte, A.R.C.; Mišan, A. Natural deep eutectic solvent (NADES) extraction improves polyphenol yield and antioxidant activity of wild thyme (Thymus serpyllum L.) extracts. Molecules 2022, 27, 1508. [Google Scholar] [CrossRef] [PubMed]
- Makris, D.P.; Lalas, S. Glycerol and glycerol-based deep eutectic mixtures as emerging green solvents for polyphenol extraction: The evidence so far. Molecules 2020, 25, 5842. [Google Scholar] [CrossRef]
- Mendes, M.A.; Souza, B.M.d.; Marques, M.R.; Palma, M.S. The effect of glycerol on signal supression during electrospray ionization analysis of proteins. Spectroscopy 2004, 18, 339–345. [Google Scholar] [CrossRef]
- Müller, E.-C. Mass Spectrometry: ESI. In Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1020–1022. [Google Scholar] [CrossRef]
- Shalliker, R.A.; Guiochon, G. Understanding the importance of the viscosity contrast between the sample solvent plug and the mobile phase and its potential consequence in two-dimensional high-performance liquid chromatography. J. Chromatogr. A 2009, 1216, 787–793. [Google Scholar] [CrossRef]
- Canton, M.; Hubert, J.; Poigny, S.; Roe, R.; Brunel, Y.; Nuzillard, J.-M.; Renault, J.-H. Dereplication of natural extracts diluted in glycerin: Physical suppression of glycerin by Centrifugal Partition Chromatography combined with presaturation of solvent signals in 13C-Nuclear Magnetic Resonance spectroscopy. Molecules 2020, 25, 5061. [Google Scholar] [CrossRef]
- Berecry, R. Production of the high anthocyanin plum cultivar,‘Queen Garnet’, as a new ingredient for the functional food market. In Proceedings of the XXIX International Horticultural Congress on Horticulture: Sustaining Lives, Livelihoods and Landscapes (IHC2014), Brisbane, Australia, 17 August 2014; Volume 1120, p. 523. [Google Scholar]
- Kodagoda, G.; Hong, H.T.; O’Hare, T.J.; Sultanbawa, Y.; Topp, B.; Netzel, M.E. Effect of Storage on the Nutritional Quality of Queen Garnet Plum. Foods 2021, 10, 352. [Google Scholar] [CrossRef]
- Bobrich, A.; Fanning, K.J.; Rychlik, M.; Russell, D.; Topp, B.; Netzel, M. Phytochemicals in Japanese plums: Impact of maturity and bioaccessibility. Food Res. Int. 2014, 65, 20–26. [Google Scholar] [CrossRef]
- Cortez, R.; Luna-Vital, D.A.; Margulis, D.; Gonzalez de Mejia, E. Natural pigments: Stabilization methods of anthocyanins for food applications. Compr. Rev. Food Sci. Food Saf. 2017, 16, 180–198. [Google Scholar] [CrossRef] [PubMed]
- Hay, T.; Prakash, S.; Daygon, V.D.; Fitzgerald, M. Review of edible Australian flora for colour and flavour additives: Appraisal of suitability and ethicality for bushfoods as natural additives to facilitate new industry growth. Trends Food Sci. Technol. 2022, 129, 74–87. [Google Scholar] [CrossRef]
- Bouarab Chibane, L.; Degraeve, P.; Ferhout, H.; Bouajila, J.; Oulahal, N. Plant antimicrobial polyphenols as potential natural food preservatives. J. Sci. Food Agric. 2019, 99, 1457–1474. [Google Scholar] [CrossRef] [PubMed]
- Nastasi, J.R.; Kontogiorgos, V.; Daygon, V.D.; Fitzgerald, M.A. Pectin-based films and coatings with plant extracts as natural preservatives: A systematic review. Trends Food Sci. Technol. 2022, 120, 193–211. [Google Scholar] [CrossRef]
- Netzel, M.; Fanning, K.; Netzel, G.; Zabaras, D.; Karagianis, G.; Treloar, T.; Russell, D.; Stanley, R. Urinary excretion of antioxidants in healthy humans following queen garnet plum juice ingestion: A new plum variety rich in antioxidant compounds. J. Food Biochem. 2012, 36, 159–170. [Google Scholar] [CrossRef]
- Santhakumar, A.B.; Kundur, A.R.; Fanning, K.; Netzel, M.; Stanley, R.; Singh, I. Consumption of anthocyanin-rich Queen Garnet plum juice reduces platelet activation related thrombogenesis in healthy volunteers. J. Funct. Foods 2015, 12, 11–22. [Google Scholar] [CrossRef]
- Santhakumar, A.B.; Kundur, A.R.; Sabapathy, S.; Stanley, R.; Singh, I. The potential of anthocyanin-rich Queen Garnet plum juice supplementation in alleviating thrombotic risk under induced oxidative stress conditions. J. Funct. Foods 2015, 14, 747–757. [Google Scholar] [CrossRef]
- Bhaswant, M.; Brown, L.; Mathai, M.L. Queen Garnet plum juice and raspberry cordial in mildly hypertensive obese or overweight subjects: A randomized, double-blind study. J. Funct. Foods 2019, 56, 119–126. [Google Scholar] [CrossRef]
- Selamoglu, Z. Polyphenolic compounds in human health with pharmacological properties. J. Tradit. Med. Clin. Naturop. 2017, 6, e137. [Google Scholar] [CrossRef]
- Röst, H.L.; Rosenberger, G.; Navarro, P.; Gillet, L.; Miladinović, S.M.; Schubert, O.T.; Wolski, W.; Collins, B.C.; Malmström, J.; Malmström, L. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014, 32, 219–223. [Google Scholar] [CrossRef]
- Adusumilli, R.; Mallick, P. Data conversion with ProteoWizard msConvert. Proteom. Methods Protoc. 2017, 1550, 339–368. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Harrington, R.A.; Adhikari, V.; Rayner, M.; Scarborough, P. Nutrient composition databases in the age of big data: FoodDB, a comprehensive, real-time database infrastructure. BMJ Open 2019, 9, e026652. [Google Scholar] [CrossRef] [PubMed]
- de Matos, P.; Dekker, A.; Ennis, M.; Hastings, J.; Haug, K.; Turner, S.; Steinbeck, C. ChEBI: A chemistry ontology and database. J. Cheminform. 2010, 2, 1. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Telukunta, K.K.; Döring, K.; Simoben, C.V.; Moumbock, A.F.A.; Malange, Y.; Njume, L.; Yong, J.; Sippl, W.; Günther, S. NANPDB: A Resource for Natural Products from Northern African Sources. J. Nat. Prod. 2017, 80, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Shinbo, Y.; Nakamura, Y.; Altaf-Ul-Amin, M.; Asahi, H.; Kurokawa, K.; Arita, M.; Saito, K.; Ohta, D.; Shibata, D.; Kanaya, S. KNApSAcK: A comprehensive species-metabolite relationship database. In Plant Metabolomics; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–181. [Google Scholar]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of open natural products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis: Chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Christou, A.; Stavrou, I.J.; Kapnissi-Christodoulou, C.P. Continuous and pulsed ultrasound-assisted extraction of carob’s antioxidants: Processing parameters optimization and identification of polyphenolic composition. Ultrason. Sonochem. 2021, 76, 105630. [Google Scholar] [CrossRef] [PubMed]
- Welton, T.; Reichardt, C. Solvents and Solvent Effects in Organic Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- El Kantar, S.; Rajha, H.N.; Boussetta, N.; Vorobiev, E.; Maroun, R.G.; Louka, N. Green extraction of polyphenols from grapefruit peels using high voltage electrical discharges, deep eutectic solvents and aqueous glycerol. Food Chem. 2019, 295, 165–171. [Google Scholar] [CrossRef]
- Amin, R.; Ahmed, D.; Aydar, A.Y.; Qamar, M.T. Modelling of polyphenol and flavonoid extraction from bottle gourd fruit using green and cost effective LTTM glycerol-ammonium acetate in neat and diluted forms. J. Food Meas. Charact. 2022, 16, 3372–3384. [Google Scholar] [CrossRef]
- Jamshaid, S.; Ahmed, D.; Aydar, A.Y. Ultrasound-assisted extraction optimization of polyphenols, flavonoids, and antioxidant compounds from fruit of Melia azedarach using a glycerol-based green deep eutectic solvent. J. Food Process. Preserv. 2022, 46, e16657. [Google Scholar] [CrossRef]
- Jamshaid, S.; Ahmed, D. Optimization of ultrasound-assisted extraction of valuable compounds from fruit of Melia azedarach with glycerol-choline chloride deep eutectic solvent. Sustain. Chem. Pharm. 2022, 29, 100827. [Google Scholar] [CrossRef]
- Kowalska, G.; Wyrostek, J.; Kowalski, R.; Pankiewicz, U. Evaluation of glycerol usage for the extraction of anthocyanins from black chokeberry and elderberry fruits. J. Appl. Res. Med. Aromat. Plants 2021, 22, 100296. [Google Scholar] [CrossRef]
- Philippi, K.; Tsamandouras, N.; Grigorakis, S.; Makris, D.P. Ultrasound-assisted green extraction of eggplant peel (Solanum melongena) polyphenols using aqueous mixtures of glycerol and ethanol: Optimisation and kinetics. Environ. Process. 2016, 3, 369–386. [Google Scholar] [CrossRef]
- Abdoun, R.; Grigorakis, S.; Kellil, A.; Loupassaki, S.; Makris, D.P. Process optimization and stability of waste orange peel polyphenols in extracts obtained with organosolv thermal treatment using glycerol-based solvents. ChemEngineering 2022, 6, 35. [Google Scholar] [CrossRef]
- Makris, D.P.; Passalidi, V.; Kallithraka, S.; Mourtzinos, I. Optimization of polyphenol extraction from red grape pomace using aqueous glycerol/tartaric acid mixtures and response surface methodology. Prep. Biochem. Biotechnol. 2016, 46, 176–182. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Palaiologou, E.; Papadakis, E.N.; Makris, D.P.; Biliaderis, C.G.; Mourtzinos, I. Insights on the impact of deep eutectic solvents on the composition of the extracts from lemon (Citrus limon L.) peels analyzed by a novel RP-LC–QTOF-MS/MS method. Eur. Food Res. Technol. 2022, 248, 2913–2927. [Google Scholar] [CrossRef]
- Panchal, S.K.; Brown, L. Tropical fruits from Australia as potential treatments for metabolic syndrome. Curr. Opin. Pharmacol. 2022, 63, 102182. [Google Scholar] [CrossRef] [PubMed]
- Ahn-Jarvis, J.H.; Parihar, A.; Doseff, A.I. Dietary flavonoids for immunoregulation and cancer: Food design for targeting disease. Antioxidants 2019, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Lanuza, F.; Bondonno, N.P.; Zamora-Ros, R.; Rostgaard-Hansen, A.L.; Tjønneland, A.; Landberg, R.; Halkjær, J.; Andres-Lacueva, C. Comparison of Flavonoid Intake Assessment Methods Using USDA and Phenol Explorer Databases: Subcohort Diet, Cancer and Health-Next Generations—MAX Study. Front. Nutr. 2022, 9, 873774. [Google Scholar] [CrossRef] [PubMed]
- Juszczak, A.M.; Marijan, M.; Jakupović, L.; Tomczykowa, M.; Tomczyk, M.; Zovko Končić, M. Glycerol and Natural Deep Eutectic Solvents Extraction for Preparation of Luteolin-Rich Jasione montana Extracts with Cosmeceutical Activity. Metabolites 2023, 13, 32. [Google Scholar] [CrossRef]
- Manousaki, A.; Jancheva, M.; Grigorakis, S.; Makris, D.P. Extraction of antioxidant phenolics from agri-food waste biomass using a newly designed glycerol-based natural low-transition temperature mixture: A comparison with conventional eco-friendly solvents. Recycling 2016, 1, 194. [Google Scholar] [CrossRef]






| Step | SPE Fractions | Volume (mL) | Glycerol Content (mg mL−1) | |
|---|---|---|---|---|
| GW | GEW | |||
| 1 | Load | 2 | 705.4 ± 6.82 | 492.65 ± 9.95 |
| 2 | Wash H2O | 1 | 33.63 ± 5.2 | 22.77 ± 10.75 |
| 3 | Wash H2O | 1 | 2.73 ± 0.89 | 2.16 ± 2.02 |
| 4 | Elute MeOH | 3 | 0.52 ± 0.11 | 0.48 ± 0.05 |
| 742.29 ± 8.3 | 520.73 ± 5.42 | |||
| Glycerol content before SPE (mg mL−1): | 744.21 ± 1.1 | 525.25 ± 0.9 | ||
| Recovery (%): | 99.7% | 99.1% | ||
| RT (min) | Adduct | Putative Metabolite Name | Metabolite Class | Molecular Formula | ES Theoretical m/z | ES Found m/z | m/z Error (Da) | MS/MS ES (+)/(−) Fragments | References (ID) | ID (1–4) |
|---|---|---|---|---|---|---|---|---|---|---|
| 8.85 | [M+] + | Cyanidin-3-O-glucoside | Anthocyanidin-3-O- glycosides | C21H21O11 | 449.101 | 449.106 | 0.002 | 449.106 (M+), 287.051 (M+ − Glu) | standard | 1 |
| 9.53 | [M+] + | Cyanidin-3-O-rutinoside | Anthocyanidin-3-O- glycosides | C27H31O15 | 595.161 | 595.163 | −0.017 | 595.161 (M+), 449.109 (M + − Rha), 287.054 (M + − Rha − Glu) | 441674 | 2 |
| 13.13 | [M + H] + | Quercetin-3-O-rutinoside | Flavonoid-3-O- glycosides | C27H30O16 | 611.155 | 610.153 | −0.017 | 611.155 (M + H), 465.101 (M + H − Rha), 303.054 (M + H − Rha − Glu) | standard | 1 |
| 13.18 | [M + H] + | Quercetin-3-O-glucoside | Flavonoid-3-O- glycosides | C21H20O12 | 465.097 | 464.095 | 0.006 | 465.097 (M + H), 303.052 (M + H − Glu) | 5280804 | 2 |
| 14.06 | [M − H] − | Quercetin-3-O-xyloside | Flavonoid-3-O- glycosides | C20H18O11 | 433.075 | 434.085 | 0.003 | 433.075 (M − H), 301.019 (M − H − Xyl) | 5321278 | 2 |
| 13.27 | [M + H] + | Quercetin-3-O-rhamnoside | Flavonoid-3-O- glycosides | C27H30O16 | 449.104 | 448.101 | 0.004 | 449.104 (M + H), 303.052 (M + H − Rha) | 5280459 | 2 |
| 13.2 | [M + H] + | Quercetin | Flavonol | C15H10O7 | 303.047 | 302.043 | 0.003 | 303.047 (M + H) 257.044 (M + H − C2H2O2), 229.049 (M + H − CO), 201.054 (M + H − CO), 153.018 (M + H − C5H4) | standard | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nastasi, J.R.; Daygon, V.D.; Kontogiorgos, V.; Fitzgerald, M.A. Qualitative Analysis of Polyphenols in Glycerol Plant Extracts Using Untargeted Metabolomics. Metabolites 2023, 13, 566. https://doi.org/10.3390/metabo13040566
Nastasi JR, Daygon VD, Kontogiorgos V, Fitzgerald MA. Qualitative Analysis of Polyphenols in Glycerol Plant Extracts Using Untargeted Metabolomics. Metabolites. 2023; 13(4):566. https://doi.org/10.3390/metabo13040566
Chicago/Turabian StyleNastasi, Joseph Robert, Venea Dara Daygon, Vassilis Kontogiorgos, and Melissa A. Fitzgerald. 2023. "Qualitative Analysis of Polyphenols in Glycerol Plant Extracts Using Untargeted Metabolomics" Metabolites 13, no. 4: 566. https://doi.org/10.3390/metabo13040566