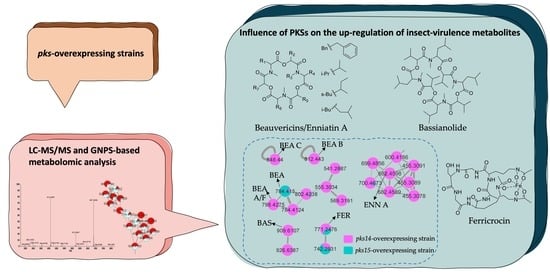
Metabolomic Analysis Demonstrates the Impacts of Polyketide Synthases PKS14 and PKS15 on the Production of Beauvericins, Bassianolide, Enniatin A, and Ferricrocin in Entomopathogen Beauveria bassiana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Generating Overexpressing Strains of pks14 and pks15
2.3. Phenotypic Characterization of pks14- and pks15-Overexpressing Strains
2.4. Metabolomic Preparation of OEpks14, OEpks15, Δpks14, and Δpks15 from Culture and In Vivo Samples
2.5. Metabolomic Analysis Using LC-MS and LC-MS/MS
2.6. Molecular Networking, Chemical Classification, and Structural Elucidation
2.6.1. LC-MS/MS Data Processing
2.6.2. Molecular Networking
3. Results
3.1. pks14- and pks15-Overexpressing Strains Exhibit Increased Insect Virulence
3.2. pks14 and pks15 Strains Differentially Express a Number of Metabolites, Including Insect Virulence Factors
3.3. Insect Virulence Factors Were Found Mainly for OEpks14 in Culture
3.4. Beauvericins and Ferricrocin Were Upregulated in OEpks15 In Vivo
3.5. pks14 Overexpression in Culture and pks15 Overexpression In Vivo Strongly Stimulated Insect-Virulence Metabolite Production
3.6. pks14 and pks15 Promoters Share Motifs with Beauvericin and Bassianolide Gene Cluster Promoters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wraight, S.P.; Ramos, M.E.; Avery, P.B.; Jaronski, S.T.; Vandenberg, J.D. Comparative virulence of Beauveria bassiana isolates against lepidopteran pests of vegetable crops. J. Invertebr. Pathol. 2010, 103, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Xu, F.; Zi, J.; Wang, S.; Gage, D.; Zeng, J.; Zhan, J. Engineered production of fungal anticancer cyclooligomer depsipeptides in Saccharomyces cerevisiae. Metab. Eng. 2013, 18, 60–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Xu, L. Beauvericin, a bioactive compound produced by fungi: A short review. Molecules 2012, 17, 2367–2377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jirakkakul, J.; Cheevadhanarak, S.; Punya, J.; Chutrakul, C.; Senachak, J.; Buajarern, T.; Tanticharoen, M.; Amnuaykanjanasin, A. Tenellin acts as an iron chelator to prevent iron-generated reactive oxygen species toxicity in the entomopathogenic fungus Beauveria bassiana. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Feng, P.; Shang, Y.; Cen, K.; Wang, C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 11365–11370. [Google Scholar] [CrossRef] [Green Version]
- Sy-Cordero, A.A.; Pearce, C.J.; Oberlies, N.H. Revisiting the enniatins: A review of their isolation, biosynthesis, structure determination and biological activities. J. Antibiot. 2012, 65, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Orozco, R.; Wijeratne, E.M.; Gunatilaka, A.A.; Stock, S.P.; Molnar, I. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem. Biol. 2008, 15, 898–907. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Orozco, R.; Kithsiri Wijeratne, E.M.; Espinosa-Artiles, P.; Leslie Gunatilaka, A.A.; Patricia Stock, S.; Molnar, I. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet. Biol. 2009, 46, 353–364. [Google Scholar] [CrossRef]
- Wallner, A.; Blatzer, M.; Schrettl, M.; Sarg, B.; Lindner, H.; Haas, H. Ferricrocin, a siderophore involved in intra-and transcellular iron distribution in Aspergillus fumigatus. Appl. Environ. Microbiol. 2009, 75, 4194–4196. [Google Scholar] [CrossRef] [Green Version]
- Eisendle, M.; Schrettl, M.; Kragl, C.; Müller, D.; Illmer, P.; Haas, H. The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination, and sexual development in Aspergillus nidulans. Eukaryot. Cell 2006, 5, 1596–1603. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.M.; Donzelli, B.G.; Krasnoff, S.B.; Keyhani, N.O. Discovering the secondary metabolite potential encoded within entomopathogenic fungi. Nat. Prod. Rep. 2014, 31, 1287–1305. [Google Scholar] [CrossRef] [PubMed]
- Hof, C.; Eisfeld, K.; Welzel, K.; Antelo, L.; Foster, A.J.; Anke, H. Ferricrocin synthesis in Magnaporthe grisea and its role in pathogenicity in rice. Mol. Plant Pathol. 2007, 8, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Jirakkakul, J.; Wichienchote, N.; Likhitrattanapisal, S.; Ingsriswang, S.; Yoocha, T.; Tangphatsornruang, S.; Wasuwan, R.; Cheevadhanarak, S.; Tanticharoen, M.; Amnuaykanjanasin, A. Iron homeostasis in the absence of ferricrocin and its consequences in fungal development and insect virulence in Beauveria bassiana. Sci. Rep. 2021, 11, 19624. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Sabiha, Y.; Loss, O.; Eisendle, M.; Wallner, A.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007, 3, 1195–1207. [Google Scholar] [CrossRef]
- Giuliano Garisto Donzelli, B.; Gibson, D.M.; Krasnoff, S.B. Intracellular siderophore but not extracellular siderophore is required for full virulence in Metarhizium robertsii. Fungal Genet. Biol. 2015, 82, 56–68. [Google Scholar] [CrossRef]
- Toopaang, W.; Bunnak, W.; Srisuksam, C.; Wattananukit, W.; Tanticharoen, M.; Yang, Y.-L.; Amnuaykanjanasin, A. Microbial polyketides and their roles in insect virulence: From genomics to biological functions. Nat. Prod. Rep. 2022, 39, 2008–2029. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Jin, K.; Ying, S.H.; Zhang, Y.; Xiao, G.; Shang, Y.; Duan, Z.; Hu, X.; Xie, X.Q.; Zhou, G.; et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar] [CrossRef] [Green Version]
- Punya, J.; Swangmaneecharern, P.; Pinsupa, S.; Nitistaporn, P.; Phonghanpot, S.; Kunathigan, V.; Cheevadhanarak, S.; Tanticharoen, M.; Amnuaykanjanasin, A. Phylogeny of type I polyketide synthases (PKSs) in fungal entomopathogens and expression analysis of PKS genes in Beauveria bassiana BCC 2660. Fungal Biol. 2015, 119, 538–550. [Google Scholar] [CrossRef]
- Srisuksam, C.; Punya, J.; Wattanachaisaereekul, S.; Toopaang, W.; Cheevadhanarak, S.; Tanticharoen, M.; Amnuaykanjanasin, A. The reducing clade IIb polyketide synthase PKS14 acts as a virulence determinant of the entomopathogenic fungus Beauveria bassiana. FEMS Microbiol. Lett. 2018, 365, 1578–1586. [Google Scholar] [CrossRef]
- Toopaang, W.; Phonghanpot, S.; Punya, J.; Panyasiri, C.; Klamchao, K.; Wasuwan, R.; Srisuksam, C.; Sangsrakru, D.; Sonthirod, C.; Tangphatsornruang, S.; et al. Targeted disruption of the polyketide synthase gene pks15 affects virulence against insects and phagocytic survival in the fungus Beauveria bassiana. Fungal Biol. 2017, 121, 664–675. [Google Scholar] [CrossRef]
- Udompaisarn, S.; Toopaang, W.; Sae-Ueng, U.; Srisuksam, C.; Wichienchote, N.; Wasuwan, R.; Nahar, N.A.S.; Tanticharoen, M.; Amnuaykanjanasin, A. The polyketide synthase PKS15 has a crucial role in cell wall formation in Beauveria bassiana. Sci. Rep. 2020, 10, 12630. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, 1743–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, E.; da Graça, J.P.; Porto, C.; Martin do Prado, R.; Hoffmann-Campo, C.B.; Meyer, M.C.; de Oliveira Nunes, E.; Pilau, E.J. Unraveling Asian Soybean Rust metabolomics using mass spectrometry and Molecular Networking approach. Sci. Rep. 2020, 10, 138. [Google Scholar] [CrossRef] [Green Version]
- Purves, K.; Macintyre, L.; Brennan, D.; Hreggviðsson, G.Ó.; Kuttner, E.; Ásgeirsdóttir, M.E.; Young, L.C.; Green, D.H.; Edrada-Ebel, R.; Duncan, K.R. Using molecular networking for microbial secondary metabolite bioprospecting. Metabolites 2016, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DesRochers, N.; Walsh, J.P.; Renaud, J.B.; Seifert, K.A.; Yeung, K.K.; Sumarah, M.W. Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng. Metabolites 2020, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anjard, C.; Su, Y.; Loomis, W.F. The polyketide MPBD initiates the SDF-1 signaling cascade that coordinates terminal differentiation in Dictyostelium. Eukaryot. Cell 2011, 10, 956–963. [Google Scholar] [CrossRef] [Green Version]
- Narita, T.B.; Chen, Z.-h.; Schaap, P.; Saito, T. The hybrid type polyketide synthase SteelyA is required for cAMP signalling in early Dictyostelium development. PLoS ONE 2014, 9, e106634. [Google Scholar] [CrossRef]
- Amnuaykanjanasin, A.; Epstein, L. A class Vb chitin synthase in Colletotrichum graminicola is localized in the growing tips of multiple cell types, in nascent septa, and during septum conversion to an end wall after hyphal breakage. Protoplasma 2006, 227, 155–164. [Google Scholar] [CrossRef]
- Epstein, L.; Lusnak, K.; Kaur, S. Transformation-Mediated Developmental Mutants of Glomerella graminicola (Colletotrichum graminicola). Fungal Genet. Biol. 1998, 23, 189–203. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Silao, F.G.S.; Bigol, U.G.; Bungay, A.A.C.; Nicolas, M.G.; Heitman, J.; Chen, Y.-L. Calcineurin is required for pseudohyphal growth, virulence, and drug resistance in Candida lusitaniae. PLoS ONE 2012, 7, e44192. [Google Scholar] [CrossRef] [Green Version]
- de Llanos, R.; Fernández-Espinar, M.T.; Querol, A. A comparison of clinical and food Saccharomyces cerevisiae isolates on the basis of potential virulence factors. Antonie Van Leeuwenhoek 2006, 90, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Kharasch, E.D.; Mitchell, D.; Coles, R. Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J. Clin. Pharmacol. 2008, 48, 464–474. [Google Scholar] [CrossRef]
- Masters, A.R.; Gufford, B.T.; Lu, J.B.L.; Metzger, I.F.; Jones, D.R.; Desta, Z. Chiral plasma pharmacokinetics and urinary excretion of bupropion and metabolites in healthy volunteers. J. Pharmacol. Exp. Ther. 2016, 358, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Sager, J.E.; Tripathy, S.; Price, L.S.; Nath, A.; Chang, J.; Stephenson-Famy, A.; Isoherranen, N. In vitro to in vivo extrapolation of the complex drug-drug interaction of bupropion and its metabolites with CYP2D6; simultaneous reversible inhibition and CYP2D6 downregulation. Biochem. Pharmacol. 2017, 123, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, S.; Funk, A.N.; Scherlach, K.; Schroeckh, V.; Shelest, E.; Horn, U.; Hertweck, C.; Brakhage, A.A. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol. 2010, 76, 8143–8149. [Google Scholar] [CrossRef] [Green Version]
- Wiemann, P.; Guo, C.-J.; Palmer, J.M.; Sekonyela, R.; Wang, C.C.C.; Keller, N.P. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl. Acad. Sci. USA 2013, 110, 17065–17070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, R.; Chhabra, A.; Phatale, P.A.; Samrat, S.K.; Sharma, J.; Gosain, A.; Mohanty, D.; Saran, S.; Gokhale, R.S. Dissecting the functional role of polyketide synthases in Dictyostelium discoideum: Biosynthesis of the differentiation regulating factor 4-methyl-5-pentylbenzene-1,3-diol. J. Biol. Chem. 2008, 283, 11348–11354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narita, T.B.; Koide, K.; Morita, N.; Saito, T. Dictyostelium hybrid polyketide synthase, SteelyA, produces 4-methyl-5-pentylbenzene-1, 3-diol and induces spore maturation. FEMS Microbiol. Lett. 2011, 319, 82–87. [Google Scholar] [CrossRef] [Green Version]
- Maor, U.; Barda, O.; Sadhasivam, S.; Bi, Y.; Levin, E.; Zakin, V.; Prusky, D.B.; Sionov, E. Functional roles of LaeA, polyketide synthase, and glucose oxidase in the regulation of ochratoxin A biosynthesis and virulence in Aspergillus carbonarius. Mol. Plant Pathol. 2021, 22, 117–129. [Google Scholar] [CrossRef] [PubMed]







| Compound | Chemical Formula | Theoretical m/z | Observed m/z | Adduct | RT (min) | m/z Error (ppm) | MS/MS Fragment |
|---|---|---|---|---|---|---|---|
| Enniatin A | C36H63N3O9 | 682.4643 | 682.4590 | [M+H]+ | 6.03 | −7.76 | 250.1410, 350.1932, 477.2928, 577.3452 |
| Ferricrocin | C28H44FeN9O13 | 771.2486 | 771.2446 | [M+H]+ | 1.35 | −5.19 | 455.1114, 524.1331, 599.1656 |
| Beauvericin | C45H57N3O9 | 784.4173 | 784.4120 | [M+H]+ | 5.85 | −6.75 | 262.1443, 362.1956, 523.2801, 623. 3325 |
| Beauvericin A/F | C46H59N3O9 | 798.4332 | 798.4277 | [M+H]+ | 6.00 | −6.89 | 262.1443, 376.2125, 537.2969, 637.3496 |
| Beauvericin B | C47H61N3O9 | 812.4486 | 812.4425 | [M+H]+ | 6.15 | −7.50 | 398.1926, 559.2763, 673.3438 |
| Beauvericin C | C48H63N3O9 | 848.4462 | 848.4398 | [M+Na]+ | 6.42 | −7.54 | 412.2090, 573.2927, 687.3608 |
| Bassianolide | C48H84N4O12 | 909.6164 | 909.6098 | [M+H]+ | 6.43 | −7.26 | 456.3088, 555.3586, 682.4649, 782.5096 |
| Compound | Chemical Formula | Theoretical m/z | Observed m/z | Adduct | RT (min) | m/z Error (ppm) | MS/MS Fragment |
|---|---|---|---|---|---|---|---|
| Ferricrocin | C28H44FeN9O13 | 771.2486 | 771.2498 | [M+H]+ | 1.35 | 1.55 | 455.1108, 542.1455, 599.1639 |
| Beauvericin | C45H57N3O9 | 784.4173 | 784.4157 | [M+H]+ | 5.86 | −2.04 | 262.1454, 362.1982, 523.2820, 623.3319 |
| Beauvericin A/ F | C46H59N3O9 | 798.4332 | 798.4305 | [M+H]+ | 6.14 | −3.38 | 398.1949, 559.2782, 659.3314 |
| Beauvericin B | C47H61N3O9 | 812.4486 | 812.4464 | [M+H]+ | 6.34 | −2.70 | 537.2957, 651.3608 |
| Beauvericin C | C48H63N3O9 | 826.4643 | 826.4626 | [M+H]+ | 6.50 | −2.05 | 276.1596, 390.2290, 551.3116, 665.3816 |
| Beauvericin D | C44H55N3O9 | 770.4017 | 770.4002 | [M+H]+ | 5.85 | −1.94 | 531.2462, 631.2984 |
| Bassanolide | C48H84N4O12 | 931.5983 | 931.5959 | [M+Na]+ | 6.42 | −2.58 | 447.2940, 577.3474, 704.4473, 804.5002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toopaang, W.; Panyawicha, K.; Srisuksam, C.; Hsu, W.-C.; Lin, C.-C.; Tanticharoen, M.; Yang, Y.-L.; Amnuaykanjanasin, A. Metabolomic Analysis Demonstrates the Impacts of Polyketide Synthases PKS14 and PKS15 on the Production of Beauvericins, Bassianolide, Enniatin A, and Ferricrocin in Entomopathogen Beauveria bassiana. Metabolites 2023, 13, 425. https://doi.org/10.3390/metabo13030425
Toopaang W, Panyawicha K, Srisuksam C, Hsu W-C, Lin C-C, Tanticharoen M, Yang Y-L, Amnuaykanjanasin A. Metabolomic Analysis Demonstrates the Impacts of Polyketide Synthases PKS14 and PKS15 on the Production of Beauvericins, Bassianolide, Enniatin A, and Ferricrocin in Entomopathogen Beauveria bassiana. Metabolites. 2023; 13(3):425. https://doi.org/10.3390/metabo13030425
Chicago/Turabian StyleToopaang, Wachiraporn, Kullyanee Panyawicha, Chettida Srisuksam, Wei-Chen Hsu, Ching-Chih Lin, Morakot Tanticharoen, Yu-Liang Yang, and Alongkorn Amnuaykanjanasin. 2023. "Metabolomic Analysis Demonstrates the Impacts of Polyketide Synthases PKS14 and PKS15 on the Production of Beauvericins, Bassianolide, Enniatin A, and Ferricrocin in Entomopathogen Beauveria bassiana" Metabolites 13, no. 3: 425. https://doi.org/10.3390/metabo13030425