A Multimodal Desorption Electrospray Ionisation Workflow Enabling Visualisation of Lipids and Biologically Relevant Elements in a Single Tissue Section
Abstract
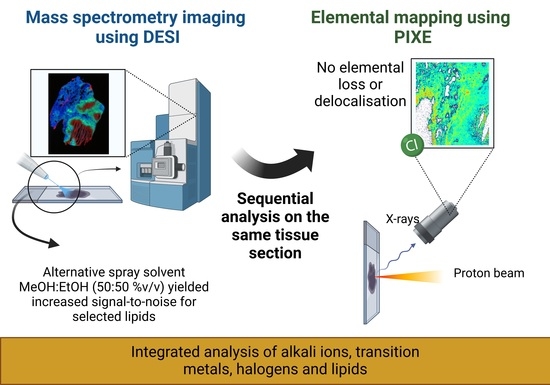
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.1.1. Homogenized Tissue
2.1.2. Snap-Frozen Lung Tissue
2.2. DESI Imaging
Data Analysis—DESI
2.3. Ion Beam Analysis
Data Analysis—Ion Beam Analysis
3. Results
3.1. Homogenized Tissue
3.2. Snap-Frozen Lung Tissue from Rabbits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walter, A.; Paul-Gilloteaux, P.; Plochberger, B.; Sefc, L.; Verkade, P.; Mannheim, J.G.; Slezak, P.; Unterhuber, A.; Marchetti-Deschmann, M.; Ogris, M.; et al. Correlated Multimodal Imaging in Life Sciences: Expanding the Biomedical Horizon. Front. Phys. 2020, 8, 47. [Google Scholar] [CrossRef]
- Matusch, A.; Fenn, L.S.; Depboylu, C.; Klietz, M.; Strohmer, S.; McLean, J.A.; Becker, J.S. Combined Elemental and Biomolecular Mass Spectrometry Imaging for Probing the Inventory of Tissue at a Micrometer Scale. Anal. Chem. 2012, 84, 3170–3178. [Google Scholar] [CrossRef]
- Svirkova, A.; Turyanskaya, A.; Perneczky, L.; Streli, C.; Marchetti-Deschmann, M. Multimodal imaging of undecalcified tissue sections by MALDI MS and μXRF. Analyst. 2018, 143, 2587–2595. [Google Scholar] [CrossRef] [PubMed]
- Touboul, D.; Roy, S.; Germain, D.P.; Chaminade, P.; Brunelle, A.; Laprévote, O. MALDI-TOF and cluster-TOF-SIMS imaging of Fabry disease biomarkers. Int. J. Mass. Spectrom. 2007, 260, 158–165. [Google Scholar] [CrossRef]
- Flint, L.E.; Hamm, G.; Ready, J.D.; Ling, S.; Duckett, C.J.; Cross, N.A.; Cole, L.M.; Smith, D.P.; Goodwin, R.J.A.; Clench, M.R. Characterization of an Aggregated Three-Dimensional Cell Culture Model by Multimodal Mass Spectrometry Imaging. Anal. Chem. 2020, 92, 12538–12547. [Google Scholar] [CrossRef]
- Petibois, C. Imaging methods for elemental, chemical, molecular, and morphological analyses of single cells. Anal. Bioanal. Chem. 2010, 397, 2051–2065. [Google Scholar] [CrossRef]
- Perry, W.J.; Weiss, A.; Van de Plas, R.; Spraggins, J.M.; Caprioli, R.M.; Skaar, E.P. Integrated molecular imaging technologies for investigation of metals in biological systems: A brief review. Curr. Opin. Chem. Biol. 2020, 55, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Szpunar, J. Advances in analytical methodology for bioinorganic speciation analysis: Metallomics, metalloproteomics and heteroatom-tagged proteomics and metabolomics. Analyst. 2005, 130, 442–465. [Google Scholar] [CrossRef] [PubMed]
- Naga Raju, G.J.; John Charles, M.; Bhuloka Reddy, S.; Sarita, P.; Seetharami Reddy, B.; Rama Lakshmi, P.V.B.; Vijayan, V. Trace elemental analysis in cancer-afflicted tissues of penis and testis by PIXE technique. Nucl. Instrum. Methods Phys. Res. B. 2005, 229, 457–464. [Google Scholar] [CrossRef]
- Rajendran, R.; Minqin, R.; Ronald, J.A.; Rutt, B.K.; Halliwell, B.; Watt, F. Does iron inhibit calcification during atherosclerosis? Free Radic. Biol. Med. 2012, 53, 1675–1679. [Google Scholar] [CrossRef] [Green Version]
- Ren, M.Q.; Ji, X.; Vajandar, S.K.; Mi, Z.H.; Hoi, A.; Walczyk, T.; van Kan, J.A.; Bettiol, A.A.; Watt, F.; Osipowicz, T. Analytical possibilities of highly focused ion beams in biomedical field. Nucl. Instrum. Methods Phys. Res. B. 2017, 406, 15–24. [Google Scholar] [CrossRef]
- Jeynes, C.; Bailey, M.J.; Coley, H.; Kirkby, K.J.; Jeynes, C. Microbeam PIXE analysis of platinum resistant and sensitive ovarian cancer cells. Nucl. Instrum. Methods Phys. Res. B. 2010, 268, 2168–2171. [Google Scholar] [CrossRef]
- Murray, F.E.S.; Landsberg, J.P.; Williams, R.J.P.; Esiri, M.M.; Watt, F. Elemental Analysis of Neurofibrillary Tangles in Alzheimer’s Disease Using Proton-Induced X-ray Analysis. In Ciba Foundation Symposium 169-Aluminium in Biology and Medicine: Aluminium in Biology and Medicine: Ciba Foundation Symposium 169; John Wiley & Sons, Ltd.: Chichester, UK, 28 September 2007; pp. 201–216. [Google Scholar]
- Basaraba, R.J.; Bielefeldt-Ohmann, H.; Eschelbach, E.K.; Reisenhauer, C.; Tolnay, A.E.; Taraba, L.C.; Shanley, C.A.; Smith, E.A.; Bedwell, C.L.; Chlipala, E.A.; et al. Increased expression of host iron-binding proteins precedes iron accumulation and calcification of primary lung lesions in experimental tuberculosis in the guinea pig. Tuberculosis 2008, 88, 69–79. [Google Scholar] [CrossRef] [PubMed]
- González de Vega, R.; Fernández-Sánchez, M.L.; Pisonero, J.; Eiró, N.; Vizoso, F.J.; Sanz-Medel, A. Quantitative bioimaging of Ca, Fe, Cu and Zn in breast cancer tissues by LA-ICP-MS. J. Anal. At. Spectrom. 2017, 32, 671–677. [Google Scholar] [CrossRef]
- Ramos, P.; Santos, A.; Pinto, E.; Pinto, N.R.; Mendes, R.; Magalhães, T.; Almeida, A. Alkali metals levels in the human brain tissue: Anatomical region differences and age-related changes. J. Trace Elem. Med. Biol. 2016, 38, 174–182. [Google Scholar] [CrossRef]
- Salina, E.G.; Waddell, S.J.; Hoffmann, N.; Rosenkrands, I.; Butcher, P.D.; Kaprelyants, A.S. Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Shimizu, N.; Ogura, Y.; Otoki, Y.; Ito, J.; Sakaino, M.; Sano, T.; Kuwahara, S.; Takekoshi, S.; Imagi, J.; et al. Structural Analysis of Lipid Hydroperoxides Using Mass Spectrometry with Alkali Metals. J. Am. Soc. Mass Spectrom. 2021, 32, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Mavroudakis, L.; Duncan, K.D.; Lanekoff, I. Host–Guest Chemistry for Simultaneous Imaging of Endogenous Alkali Metals and Metabolites with Mass Spectrometry. Anal. Chem. 2022, 94, 2391–2398. [Google Scholar] [CrossRef]
- da Cunha, M.M.L.; Trepout, S.; Messaoudi, C.; Wu, T.-D.; Ortega, R.; Guerquin-Kern, J.-L.; Marco, S. Overview of chemical imaging methods to address biological questions. Micron. 2016, 84, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Becker, J.S. Imaging techniques for elements and element species in plant science. Metallomics. 2012, 4, 403–416. [Google Scholar] [CrossRef]
- Jeynes, C.; Colaux, J.L. Thin film depth profiling by ion beam analysis. Analyst 2016, 141, 5944–5985. [Google Scholar] [CrossRef]
- Bonta, M.; Gonzalez, J.J.; Quarles, C.D.; Russo, R.E.; Hegedus, B.; Limbeck, A. Elemental mapping of biological samples by the combined use of LIBS and LA-ICP-MS. J. Anal. At. Spectrom. 2016, 31, 252–258. [Google Scholar] [CrossRef]
- Martinez, M.; Baudelet, M. Calibration strategies for elemental analysis of biological samples by LA-ICP-MS and LIBS – A review. Anal. Bioanal. Chem. 2020, 412, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Pozebon, D.; Scheffler, G.L.; Dressler, V.L. Recent applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) for biological sample analysis: A follow-up review. J. Anal. At. Spectrom. 2017, 32, 890–919. [Google Scholar] [CrossRef]
- Pozebon, D.; Scheffler, G.L.; Dressler, V.L.; Nunes, M.A.G. Review of the applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to the analysis of biological samples. J. Anal. At. Spectrom. 2014, 29, 2204–2228. [Google Scholar] [CrossRef]
- Sussulini, A.; Becker, J.S.; Becker, J.S. Laser ablation ICP-MS: Application in biomedical research. Mass Spectrom. Rev. 2017, 36, 47–57. [Google Scholar] [CrossRef]
- Konz, I.; Fernández, B.; Fernández, M.L.; Pereiro, R.; González, H.; Álvarez, L.; Coca-Prados, M.; Sanz-Medel, A. Gold internal standard correction for elemental imaging of soft tissue sections by LA-ICP-MS: Element distribution in eye microstructures. Anal. Bioanal. Chem. 2013, 405, 3091–3096. [Google Scholar] [CrossRef]
- Fernández, B. Elemental and molecular imaging by LA-ICP-MS. Anal. Bioanal. Chem. 2019, 411, 547–548. [Google Scholar] [CrossRef] [PubMed]
- Paunesku, T.; Vogt, S.; Maser, J.; Lai, B.; Woloschak, G. X-ray fluorescence microprobe imaging in biology and medicine. J. Cell. Biochem. 2006, 99, 1489–1502. [Google Scholar] [CrossRef]
- Uo, M.; Wada, T.; Sugiyama, T. Applications of X-ray fluorescence analysis (XRF) to dental and medical specimens. Jpn. Dent. Sci. Rev. 2015, 51, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Kump, P.; Vogel-Mikuš, K. Quantification of 2D elemental distribution maps of intermediate-thick biological sections by low energy synchrotron μ-X-ray fluorescence spectrometry. J. Instrum. 2018, 13, C05014. [Google Scholar] [CrossRef]
- Collingwood, J.F.; Adams, F. Chemical imaging analysis of the brain with X-ray methods. Spectrochim. Acta Part B At. Spectrosc. 2017, 130, 101–118. [Google Scholar] [CrossRef]
- Kanngießer, B.; Malzer, W.; Pagels, M.; Lühl, L.; Weseloh, G. Three-dimensional micro-XRF under cryogenic conditions: A pilot experiment for spatially resolved trace analysis in biological specimens. Anal. Bioanal. Chem. 2007, 389, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Streli, C.; Rauwolf, M.; Turyanskaya, A.; Ingerle, D.; Wobrauschek, P. Elemental imaging of trace elements in bone samples using micro and nano-X-ray fluorescence spectrometry. Appl. Radiat. Isot. 2019, 149, 200–205. [Google Scholar] [CrossRef]
- Ishii, K.; Sugimoto, A.; Tanaka, A.; Satoh, T.; Matsuyama, S.; Yamazaki, H.; Akama, C.; Amartivan, T.; Endoh, H.; Oishi, Y.; et al. Elemental analysis of cellular samples by in-air micro-PIXE. Nucl. Instrum. Methods Phys. Res. B 2001, 181, 448–453. [Google Scholar] [CrossRef]
- Tylko, G.; Mesjasz-Przybyłowicz, J.; Przybyłowicz, W.J. In-vacuum micro-PIXE analysis of biological specimens in frozen-hydrated state. Nucl. Instrum. Methods Phys. Res. B 2007, 260, 141–148. [Google Scholar] [CrossRef]
- Vogel-Mikuš, K.; Pongrac, P.; Pelicon, P. Micro-PIXE elemental mapping for ionome studies of crop plants. Int. J. PIXE. 2014, 24, 217–233. [Google Scholar] [CrossRef]
- Ishii, K. PIXE and Its Applications to Elemental Analysis. Quantum Beam Sci. 2019, 3, 12. [Google Scholar] [CrossRef]
- Mulware, S.J. Comparative Trace Elemental Analysis in Cancerous and Noncancerous Human Tissues Using PIXE. J. Biophys. 2013, 2013, 192026. [Google Scholar] [CrossRef]
- Sakai, T.; Oikawa, M.; Sato, T.; Nagamine, T.; Moon, H.D.; Nakazato, K.; Suzuki, K. New in-air micro-PIXE system for biological applications. Nucl. Instrum. Methods Phys. Res. B. 2005, 231, 112–116. [Google Scholar] [CrossRef]
- Neumann, E.K.; Djambazova, K.V.; Caprioli, R.M.; Spraggins, J.M. Multimodal Imaging Mass Spectrometry: Next Generation Molecular Mapping in Biology and Medicine. J. Am. Soc. Mass Spectrom. 2020, 31, 2401–2415. [Google Scholar] [CrossRef]
- Buchberger, A.R.; DeLaney, K.; Johnson, J.; Li, L. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal Chem. 2018, 90, 240–265. [Google Scholar] [CrossRef]
- Dilmetz, B.A.; Lee, Y.-R.; Condina, M.R.; Briggs, M.; Young, C.; Desire, C.T.; Klingler-Hoffmann, M.; Hoffmann, P. Novel technical developments in mass spectrometry imaging in 2020: A mini review. Anal. Sci. Adv. 2021, 2, 225–237. [Google Scholar] [CrossRef]
- Eberlin, L.S. DESI-MS Imaging of Lipids and Metabolites from Biological Samples. In Mass Spectrometry in Metabolomics: Methods and Protocols; Raftery, D., Ed.; Springer: New York, NY, USA, 2014; pp. 299–311. [Google Scholar]
- Eberlin, L.S.; Ferreira, C.R.; Dill, A.L.; Ifa, D.R.; Cooks, R.G. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2011, 1811, 946–960. [Google Scholar] [CrossRef]
- Wiseman, J.M.; Ifa, D.R.; Song, Q.; Cooks, R.G. Tissue Imaging at Atmospheric Pressure Using Desorption Electrospray Ionization (DESI) Mass Spectrometry. Angew. Chem., Int. Ed. Engl. 2006, 45, 7188–7192. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.V.; Gamage, C.M.; Fernández, F.M. Imaging of Biological Tissues by Desorption Electrospray Ionization Mass Spectrometry. J. Vis. Exp. 2013, 77, e50575. [Google Scholar]
- Strittmatter, N.; Moss, J.I.; Race, A.M.; Sutton, D.; Canales, J.R.; Ling, S.; Wong, E.; Wilson, J.; Smith, A.; Howes, C.; et al. Multi-modal molecular imaging maps the correlation between tumor microenvironments and nanomedicine distribution. Theranostics 2022, 12, 2162–2174. [Google Scholar] [CrossRef]
- Lee, P.Y.; Yeoh, Y.; Omar, N.; Pung, Y.-F.; Lim, L.C.; Low, T.Y. Molecular tissue profiling by MALDI imaging: Recent progress and applications in cancer research. Crit. Rev. Clin. Lab Sci. 2021, 58, 513–529. [Google Scholar] [CrossRef]
- Harris, A.; Roseborough, A.; Mor, R.; Yeung, K.K.C.; Whitehead, S.N. Ganglioside Detection from Formalin-Fixed Human Brain Tissue Utilizing MALDI Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 479–487. [Google Scholar] [CrossRef]
- Hermann, J.; Noels, H.; Theelen, W.; Lellig, M.; Orth-Alampour, S.; Boor, P.; Jankowski, V.; Jankowski, J. Sample preparation of formalin-fixed paraffin-embedded tissue sections for MALDI-mass spectrometry imaging. Anal. Bioanal. Chem. 2020, 412, 1263–1275. [Google Scholar] [CrossRef]
- Bowman, A.P.; Bogie, J.F.J.; Hendriks, J.J.A.; Haidar, M.; Belov, M.; Heeren, R.M.A.; Ellis, S.R. Evaluation of lipid coverage and high spatial resolution MALDI-imaging capabilities of oversampling combined with laser post-ionisation. Anal. Bioanal. Chem. 2020, 412, 2277–2289. [Google Scholar] [CrossRef] [Green Version]
- Sämfors, S.; Fletcher, J.S. Lipid Diversity in Cells and Tissue Using Imaging SIMS. Annu. Rev. Anal. Chem. 2020, 13, 249–271. [Google Scholar] [CrossRef] [PubMed]
- Van Nuffel, S.; Quatredeniers, M.; Pirkl, A.; Zakel, J.; Le Caer, J.-P.; Elie, N.; Vanbellingen, Q.P.; Dumas, S.J.; Nakhleh, M.K.; Ghigna, M.-R.; et al. Multimodal Imaging Mass Spectrometry to Identify Markers of Pulmonary Arterial Hypertension in Human Lung Tissue Using MALDI-ToF, ToF-SIMS, and Hybrid SIMS. Anal. Chem. 2020, 92, 12079–12087. [Google Scholar] [CrossRef] [PubMed]
- Agüi-Gonzalez, P.; Jähne, S.; Phan, N.T.N. SIMS imaging in neurobiology and cell biology. J. Anal. At. Spectrom. 2019, 34, 1355–1368. [Google Scholar] [CrossRef]
- Sämfors, S.; Ståhlman, M.; Klevstig, M.; Borén, J.; Fletcher, J.S. Localised lipid accumulation detected in infarcted mouse heart tissue using ToF-SIMS. Int. J. Mass spectrom. 2019, 437, 77–86. [Google Scholar] [CrossRef]
- Massonnet, P.; Heeren, R.M.A. A concise tutorial review of TOF-SIMS based molecular and cellular imaging. J. Anal. At. Spectrom. 2019, 34, 2217–2228. [Google Scholar] [CrossRef]
- Illes-Toth, E.; Hale, O.J.; Hughes, J.W.; Strittmatter, N.; Rose, J.; Clayton, B.; Sargeant, R.; Jones, S.; Dannhorn, A.; Goodwin, R.J.A.; et al. Mass Spectrometry Detection and Imaging of a Non-Covalent Protein–Drug Complex in Tissue from Orally Dosed Rats. Angew. Chem. Int. Ed. Engl. 2022, 61, e202202075. [Google Scholar] [PubMed]
- Qi, K.; Wu, L.; Liu, C.; Pan, Y. Recent Advances of Ambient Mass Spectrometry Imaging and Its Applications in Lipid and Metabolite Analysis. Metabolites 2021, 11, 780. [Google Scholar] [CrossRef]
- Unsihuay, D.; Sanchez, D.M.; Laskin, J. Quantitative Mass Spectrometry Imaging of Biological Systems. Annu. Rev. Phys. Chem. 2021, 72, 307–329. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.L.; Lombi, E.; Zhao, F.-J.; Grovenor, C.R.M. Elemental imaging at the nanoscale: NanoSIMS and complementary techniques for element localisation in plants. Anal. Bioanal. Chem. 2012, 402, 3263–3273. [Google Scholar] [CrossRef]
- Liu, H.; Chen, R.; Wang, J.; Chen, S.; Xiong, C.; Wang, J.; Hou, J.; He, Q.; Zhang, N.; Nie, Z.; et al. 1,5-Diaminonaphthalene Hydrochloride Assisted Laser Desorption/Ionization Mass Spectrometry Imaging of Small Molecules in Tissues Following Focal Cerebral Ischemia. Anal. Chem. 2014, 86, 10114–10121. [Google Scholar] [CrossRef]
- de Jesus, J.M.; Costa, C.; Burton, A.; Palitsin, V.; Webb, R.; Taylor, A.; Nikula, C.; Dexter, A.; Kaya, F.; Chambers, M.; et al. Correlative Imaging of Trace Elements and Intact Molecular Species in a Single-Tissue Sample at the 50 μm Scale. Anal. Chem. 2021, 93, 13450–13458. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; de Jesus, J.; Nikula, C.; Murta, T.; Grime, G.W.; Palitsin, V.; Webb, R.; Goodwin, R.J.A.; Bunch, J.; Bailey, M.J. Exploring New Methods to Study and Moderate Proton Beam Damage for Multimodal Imaging on a Single Tissue Section. J. Am. Soc. Mass Spectrom. 2022, 33, 2263–2272. [Google Scholar] [CrossRef]
- Sans, M.; Feider, C.L.; Eberlin, L.S. Advances in mass spectrometry imaging coupled to ion mobility spectrometry for enhanced imaging of biological tissues. Curr. Opin. Chem. Biol. 2018, 42, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Lewis, H.-M.; Costa, C.; Dartois, V.; Kaya, F.; Chambers, M.; de Jesus, J.; Palitsin, V.; Webb, R.; Bailey, M.J. Colocation of Lipids, Drugs, and Metal Biomarkers Using Spatially Resolved Lipidomics with Elemental Mapping. Anal. Chem. 2022, 94, 11798–11806. [Google Scholar] [CrossRef]
- Swales, J.G.; Strittmatter, N.; Tucker, J.W.; Clench, M.R.; Webborn, P.J.H.; Goodwin, R.J.A. Spatial Quantitation of Drugs in tissues using Liquid Extraction Surface Analysis Mass Spectrometry Imaging. Sci. Rep. 2016, 6, 37648. [Google Scholar] [CrossRef]
- Subbian, S.; Tsenova, L.; Yang, G.; O’Brien, P.; Parsons, S.; Peixoto, B.; Taylor, L.; Fallows, D.; Kaplan, G. Chronic pulmonary cavitary tuberculosis in rabbits: A failed host immune response. Open Biol. 2011, 1, 110016. [Google Scholar] [CrossRef] [PubMed]
- Lamont, E.A.; Dillon, N.A.; Baughn, A.D. The Bewildering Antitubercular Action of Pyrazinamide. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, M.; Blanc, L.; Chen, P.-Y.; Dartois, V.; Prideaux, B. Spatial Quantification of Drugs in Pulmonary Tuberculosis Lesions by Laser Capture Microdissection Liquid Chromatography Mass Spectrometry (LCM-LC/MS). J. Vis. Exp. 2018, 134, e57402. [Google Scholar]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Race, A.M.; Styles, I.B.; Bunch, J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteom. 2012, 75, 5111–5112. [Google Scholar] [CrossRef] [PubMed]
- Race, A.M.; Palmer, A.D.; Dexter, A.; Steven, R.T.; Styles, I.B.; Bunch, J. SpectralAnalysis: Software for the Masses. Anal. Chem. 2016, 88, 9451–9458. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. .Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Grime, G.W.; Zeldin, O.B.; Snell, M.E.; Lowe, E.D.; Hunt, J.F.; Montelione, G.T.; Tong, L.; Snell, E.H.; Garman, E.F. High-Throughput PIXE as an Essential Quantitative Assay for Accurate Metalloprotein Structural Analysis: Development and Application. J. Am. Soc. Mass Spectrom. 2020, 142, 185–197. [Google Scholar] [CrossRef]
- Patra, M.; Salonen, E.; Terama, E.; Vattulainen, I.; Faller, R.; Lee, B.W.; Holopainen, J.; Karttunen, M. Under the Influence of Alcohol: The Effect of Ethanol and Methanol on Lipid Bilayers. Biophys. J. 2006, 90, 1121–1135. [Google Scholar] [CrossRef]
- Yin, R.; Burnum-Johnson, K.E.; Sun, X.; Dey, S.K.; Laskin, J. High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nat. Protoc. 2019, 14, 3445–3470. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, C.; De Jesus, J.; Nikula, C.; Murta, T.; Grime, G.W.; Palitsin, V.; Dartois, V.; Firat, K.; Webb, R.; Bunch, J.; et al. A Multimodal Desorption Electrospray Ionisation Workflow Enabling Visualisation of Lipids and Biologically Relevant Elements in a Single Tissue Section. Metabolites 2023, 13, 262. https://doi.org/10.3390/metabo13020262
Costa C, De Jesus J, Nikula C, Murta T, Grime GW, Palitsin V, Dartois V, Firat K, Webb R, Bunch J, et al. A Multimodal Desorption Electrospray Ionisation Workflow Enabling Visualisation of Lipids and Biologically Relevant Elements in a Single Tissue Section. Metabolites. 2023; 13(2):262. https://doi.org/10.3390/metabo13020262
Chicago/Turabian StyleCosta, Catia, Janella De Jesus, Chelsea Nikula, Teresa Murta, Geoffrey W. Grime, Vladimir Palitsin, Véronique Dartois, Kaya Firat, Roger Webb, Josephine Bunch, and et al. 2023. "A Multimodal Desorption Electrospray Ionisation Workflow Enabling Visualisation of Lipids and Biologically Relevant Elements in a Single Tissue Section" Metabolites 13, no. 2: 262. https://doi.org/10.3390/metabo13020262