Serum Metabolomics Profiling Reveals Metabolic Alterations Prior to a Diagnosis with Non-Small Cell Lung Cancer among Chinese Community Residents: A Prospective Nested Case-Control Study
Abstract
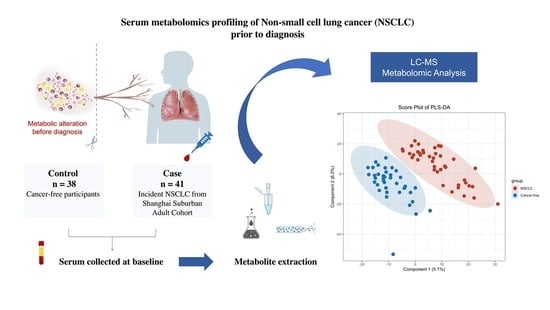
:1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Design and Subjects
2.3. Data Collection and Measurement
2.4. Metabolomics Profiling and Data Preprocessing
2.5. Statistical Analysis
2.6. Role of the Funding Sponsors
3. Results
3.1. Participant Characteristics
3.2. Significantly Changed Metabolites and Their relative Changes across NSCLC Cases and Cancer-Free Controls
3.3. Altered Metabolic Pathways across NSCLC Cases and Cancer-Free Controls
3.4. Serum Metabolite Signatures for NSCLC
3.5. Correlation between Significantly Changed Metabolites, Baseline Characteristics, and Classical Lipids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Untargeted Metabolomics Profiling
Appendix A.1.1. LC-MS Metabolites Extraction
Appendix A.1.2. LC-MS/MS Analysis
Appendix A.1.3. Quality Control Process
Appendix B


References
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Oudkerk, M.; Liu, S.; Heuvelmans, M.A.; Walter, J.E.; Field, J.K. Lung cancer LDCT screening and mortality reduction—Evidence, pitfalls and future perspectives. Nat. Rev. Clin. Oncol. 2021, 18, 135–151. [Google Scholar] [CrossRef]
- Toumazis, I.; Bastani, M.; Han, S.S.; Plevritis, S.K. Risk-Based lung cancer screening: A systematic review. Lung Cancer 2020, 147, 154–186. [Google Scholar] [CrossRef]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [PubMed]
- Schult, T.A.; Lauer, M.J.; Berker, Y.; Cardoso, M.R.; Vandergrift, L.A.; Habbel, P.; Nowak, J.; Taupitz, M.; Aryee, M.; Mino-Kenudson, M.A.; et al. Screening human lung cancer with predictive models of serum magnetic resonance spectroscopy metabolomics. Proc. Natl. Acad. Sci. USA 2021, 118, e2110633118. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nishiumi, S.; Kobayashi, K.; Shinohara, M.; Hatakeyama, Y.; Kotani, Y.; Hatano, N.; Maniwa, Y.; Nishio, W.; Bamba, T.; et al. A metabolomic approach to lung cancer. Lung Cancer 2011, 74, 284–292. [Google Scholar] [CrossRef]
- Wittmann, B.M.; Stirdivant, S.M.; Mitchell, M.W.; Wulff, J.E.; McDunn, J.E.; Li, Z.; Dennis-Barrie, A.; Neri, B.P.; Milburn, M.V.; Lotan, Y.; et al. Bladder cancer biomarker discovery using global metabolomic profiling of urine. PLoS ONE 2014, 9, e115870. [Google Scholar] [CrossRef]
- Yu, L.; Jiang, C.; Huang, S.; Gong, X.; Wang, S.; Shen, P. Analysis of urinary metabolites for breast cancer patients receiving chemotherapy by CE-MS coupled with on-line concentration. Clin. Biochem. 2013, 46, 1065–1073. [Google Scholar] [CrossRef]
- Playdon, M.C.; Ziegler, R.G.; Sampson, J.N.; Stolzenberg-Solomon, R.; Thompson, H.J.; Irwin, M.L.; Mayne, S.T.; Hoover, R.N.; Moore, S.C. Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 2017, 106, 637–649. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, B.; Wang, R.; Zhu, M.; Shao, Y.; Wang, N.; Liu, X.; Zhang, T.; Jiang, F.; Wang, W.; et al. Cohort profile: Protocol and baseline survey for the Shanghai Suburban Adult Cohort and Biobank (SSACB) study. BMJ Open 2020, 10, e035430. [Google Scholar] [CrossRef]
- The Kyoto Encyclopedia of Genes and Genomes Database. Available online: http://www.kegg.jp (accessed on 5 December 2021).
- The Human Metabolome Database. Available online: https://hmdb.ca/ (accessed on 25 December 2021).
- Merino Salvador, M.; Gómez de Cedrón, M.; Moreno Rubio, J.; Falagán Martínez, S.; Sánchez Martínez, R.; Casado, E.; Ramírez de Molina, A.; Sereno, M. Lipid metabolism and lung cancer. Crit. Rev. Oncol. Hematol. 2017, 112, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Elia, I.; Broekaert, D.; Christen, S.; Boon, R.; Radaelli, E.; Orth, M.F.; Verfaillie, C.; Grünewald, T.G.P.; Fendt, S.M. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 2017, 8, 15267. [Google Scholar] [CrossRef]
- Liu, W.; Hancock, C.N.; Fischer, J.W.; Harman, M.; Phang, J.M. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: Involvement of pyridine nucleotides. Sci. Rep. 2015, 5, 17206. [Google Scholar] [CrossRef]
- Sahu, N.; Dela Cruz, D.; Gao, M.; Sandoval, W.; Haverty, P.M.; Liu, J.; Stephan, J.P.; Haley, B.; Classon, M.; Hatzivassiliou, G.; et al. Proline Starvation Induces Unresolved ER Stress and Hinders mTORC1-Dependent Tumorigenesis. Cell Metab. 2016, 24, 753–761. [Google Scholar] [CrossRef]
- Polyak, K.; Xia, Y.; Zweier, J.L.; Kinzler, K.W.; Vogelstein, B. A model for p53-induced apoptosis. Nature 1997, 389, 300–305. [Google Scholar] [CrossRef]
- Canale, M.; Andrikou, K.; Priano, I.; Cravero, P.; Pasini, L.; Urbini, M.; Delmonte, A.; Crinò, L.; Bronte, G.; Ulivi, P. The Role of TP53 Mutations in EGFR-Mutated Non-Small-Cell Lung Cancer: Clinical Significance and Implications for Therapy. Cancers (Basel) 2022, 14, 1143. [Google Scholar] [CrossRef] [PubMed]
- Adams, E. Metabolism of proline and of hydroxyproline. Int. Rev. Connect. Tissue Res. 1970, 5, 1–91. [Google Scholar] [PubMed]
- Liu, Y.; Mao, C.; Wang, M.; Liu, N.; Ouyang, L.; Liu, S.; Tang, H.; Cao, Y.; Liu, S.; Wang, X.; et al. Cancer progression is mediated by proline catabolism in non-small cell lung cancer. Oncogene 2020, 39, 2358–2376. [Google Scholar] [CrossRef]
- Lee, K.B.; Ang, L.; Yau, W.P.; Seow, W.J. Association between Metabolites and the Risk of Lung Cancer: A Systematic Literature Review and Meta-Analysis of Observational Studies. Metabolites 2020, 10, 362. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Nikas, I.P.; Paschou, S.A.; Ryu, H.S. The Role of Nicotinamide in Cancer Chemoprevention and Therapy. Biomolecules 2020, 10, 477. [Google Scholar] [CrossRef]
- Song, S.B.; Jang, S.Y.; Kang, H.T.; Wei, B.; Jeoun, U.W.; Yoon, G.S.; Hwang, E.S. Modulation of Mitochondrial Membrane Potential and ROS Generation by Nicotinamide in a Manner Independent of SIRT1 and Mitophagy. Mol. Cells 2017, 40, 503–514. [Google Scholar] [PubMed]
- Marien, E.; Meister, M.; Muley, T.; Fieuws, S.; Bordel, S.; Derua, R.; Spraggins, J.; Van de Plas, R.; Dehairs, J.; Wouters, J.; et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int. J. Cancer 2015, 137, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Eberlin, L.S.; Gabay, M.; Fan, A.C.; Gouw, A.M.; Tibshirani, R.J.; Felsher, D.W.; Zare, R.N. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc. Natl. Acad. Sci. USA 2014, 111, 10450–10455. [Google Scholar] [CrossRef]
- Saddoughi, S.A.; Song, P.; Ogretmen, B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell. Biochem. 2008, 49, 413–440. [Google Scholar]
- Haddadi, N.; Lin, Y.; Simpson, A.M.; Nassif, N.T.; McGowan, E.M. "Dicing and Splicing" Sphingosine Kinase and Relevance to Cancer. Int. J. Mol. Sci. 2017, 18, 1891. [Google Scholar] [CrossRef]
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the sphingolipid rheostat: Evolving concepts in cancer therapy. Exp. Cell Res. 2015, 333, 195–200. [Google Scholar] [CrossRef]
- Zheng, X.; Li, W.; Ren, L.; Liu, J.; Pang, X.; Chen, X.; Kang, D.; Wang, J.; Du, G. The sphingosine kinase-1/sphingosine-1-phosphate axis in cancer: Potential target for anticancer therapy. Pharmacol. Ther. 2019, 195, 85–99. [Google Scholar] [CrossRef]
- Song, L.; Xiong, H.; Li, J.; Liao, W.; Wang, L.; Wu, J.; Li, M. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-κB pathway in human non-small cell lung cancer. Clin. Cancer Res. 2011, 17, 1839–1849. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef]
- Seow, W.J.; Shu, X.O.; Nicholson, J.K.; Holmes, E.; Walker, D.I.; Hu, W.; Cai, Q.; Gao, Y.T.; Xiang, Y.B.; Moore, S.C.; et al. Association of Untargeted Urinary Metabolomics and Lung Cancer Risk Among Never-Smoking Women in China. JAMA Netw. Open 2019, 2, e1911970. [Google Scholar] [CrossRef]
- Vandergrift, L.A.; Decelle, E.A.; Kurth, J.; Wu, S.; Fuss, T.L.; DeFeo, E.M.; Halpern, E.F.; Taupitz, M.; McDougal, W.S.; Olumi, A.F.; et al. Metabolomic Prediction of Human Prostate Cancer Aggressiveness: Magnetic Resonance Spectroscopy of Histologically Benign Tissue. Sci. Rep. 2018, 8, 4997. [Google Scholar] [CrossRef] [PubMed]
- Sonkar, K.; Ayyappan, V.; Tressler, C.M.; Adelaja, O.; Cai, R.; Cheng, M.; Glunde, K. Focus on the glycerophosphocholine pathway in choline phospholipid metabolism of cancer. NMR Biomed. 2019, 32, e4112. [Google Scholar] [CrossRef] [PubMed]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Mountford, C.E.; Wright, L.C. Organization of lipids in the plasma membranes of malignant and stimulated cells: A new model. Trends Biochem. Sci. 1988, 13, 172–177. [Google Scholar] [CrossRef]
- Cuadrado, A.; Carnero, A.; Dolfi, F.; Jiménez, B.; Lacal, J.C. Phosphorylcholine: A novel second messenger essential for mitogenic activity of growth factors. Oncogene 1993, 8, 2959–2968. [Google Scholar] [PubMed]
- Price, B.D.; Morris, J.D.; Marshall, C.J.; Hall, A. Stimulation of phosphatidylcholine hydrolysis, diacylglycerol release, and arachidonic acid production by oncogenic ras is a consequence of protein kinase C activation. J. Biol. Chem. 1989, 264, 16638–16643. [Google Scholar] [CrossRef]
- Besterman, J.M.; Duronio, V.; Cuatrecasas, P. Rapid formation of diacylglycerol from phosphatidylcholine: A pathway for generation of a second messenger. Proc. Natl. Acad. Sci. USA 1986, 83, 6785–6789. [Google Scholar] [CrossRef] [PubMed]
- Bogin, L.; Papa, M.Z.; Polak-Charcon, S.; Degani, H. TNF-induced modulations of phospholipid metabolism in human breast cancer cells. Biochim. Biophys. Acta 1998, 1392, 217–232. [Google Scholar] [CrossRef]
- Aboagye, E.O.; Bhujwalla, Z.M. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999, 59, 80–84. [Google Scholar] [PubMed]
- Galons, J.P.; Job, C.; Gillies, R.J. Increase of GPC levels in cultured mammalian cells during acidosis. A 31P MR spectroscopy study using a continuous bioreactor system. Magn. Reson. Med. 1995, 33, 422–426. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef] [PubMed]
- White-Gilbertson, S.; Mullen, T.; Senkal, C.; Lu, P.; Ogretmen, B.; Obeid, L.; Voelkel-Johnson, C. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene 2009, 28, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Siskind, L.J.; Feinstein, L.; Yu, T.; Davis, J.S.; Jones, D.; Choi, J.; Zuckerman, J.E.; Tan, W.; Hill, R.B.; Hardwick, J.M.; et al. Anti-apoptotic Bcl-2 Family Proteins Disassemble Ceramide Channels. J. Biol. Chem. 2008, 283, 6622–6630. [Google Scholar] [CrossRef]
- Blom, T.; Li, S.; Dichlberger, A.; Bäck, N.; Kim, Y.A.; Loizides-Mangold, U.; Riezman, H.; Bittman, R.; Ikonen, E. LAPTM4B facilitates late endosomal ceramide export to control cell death pathways. Nat. Chem. Biol. 2015, 11, 799–806. [Google Scholar] [CrossRef]
- Schiffmann, S.; Sandner, J.; Birod, K.; Wobst, I.; Angioni, C.; Ruckhäberle, E.; Kaufmann, M.; Ackermann, H.; Lötsch, J.; Schmidt, H.; et al. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis 2009, 30, 745–752. [Google Scholar] [CrossRef]
- Senkal, C.E.; Ponnusamy, S.; Manevich, Y.; Meyers-Needham, M.; Saddoughi, S.A.; Mukhopadyay, A.; Dent, P.; Bielawski, J.; Ogretmen, B. Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network. J. Biol. Chem. 2011, 286, 42446–42458. [Google Scholar] [CrossRef]
- Podo, F. Tumour phospholipid metabolism. NMR Biomed. 1999, 12, 413–439. [Google Scholar] [CrossRef]
- Maeda, J.; Higashiyama, M.; Imaizumi, A.; Nakayama, T.; Yamamoto, H.; Daimon, T.; Yamakado, M.; Imamura, F.; Kodama, K. Possibility of multivariate function composed of plasma amino acid profiles as a novel screening index for non-small cell lung cancer: A case control study. BMC Cancer 2010, 10, 690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, K.W.; Adkins, C.B.; Su, L.; Halpern, E.F.; Mark, E.J.; Christiani, D.C.; Cheng, L.L. Comparison of squamous cell carcinoma and adenocarcinoma of the lung by metabolomic analysis of tissue-serum pairs. Lung Cancer 2010, 68, 44–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Characteristic | NSCLC (n = 41) | Cancer-Free Control (n = 38) | Total (n = 79) | p-Value |
|---|---|---|---|---|
| Age, years | 0.371 | |||
| 41–55 | 9 (21.95%) | 8 (21.05%) | 17 (21.52%) | |
| 56–65 | 18 (43.90%) | 22 (57.89%) | 40 (50.63%) | |
| 65–75 | 14 (34.15%) | 8 (21.05%) | 22 (27.85%) | |
| Mean ± SD (years) | 61.24 ± 6.97 | 60.21 ± 6.74 | 60.75 ± 6.84 | 0.505 |
| Gender | 0.411 | |||
| Male | 21 (51.22%) | 15 (39.47%) | 36 (45.57%) | |
| Female | 20 (48.78%) | 23 (60.53%) | 43 (54.43%) | |
| Education | 0.558 | |||
| Middle school or below | 20 (48.78%) | 22 (57.89%) | 42 (53.16%) | |
| High school or above | 21 (51.22%) | 16 (42.11%) | 37 (46.84%) | |
| History of Respiratory Diseases | 0.241 | |||
| Yes | 3 (7.32%) | 0 (0.00%) | 3 (3.80%) | |
| No | 38 (92.68%) | 38 (100.00%) | 76 (96.20%) | |
| Smoking Status | 0.160 | |||
| Never | 27 (65.85%) | 31 (81.58%) | 58 (73.42%) | |
| Former | 1 (2.44%) | 0 (0.00%) | 1 (1.27%) | |
| Current | 13 (31.71%) | 7 (18.42%) | 20 (25.32%) | |
| Second-hand Exposure | 0.026 | |||
| Yes | 6 (14.63%) | 0 (0.00%) | 6 (7.59%) | |
| No | 35 (85.37%) | 38 (100.00%) | 73 (92.41%) | |
| Alcohol drinking | 0.228 | |||
| Yes | 0 (0.00%) | 2 (5.26%) | 2 (2.53%) | |
| No | 41 (100.00%) | 36 (94.74%) | 77 (97.47%) | |
| Exercise | 0.185 | |||
| Yes | 14 (34.15%) | 7 (18.42%) | 21 (26.58%) | |
| No | 27 (65.85%) | 31 (81.58%) | 58 (73.42%) | |
| BMI, kg/m2 | 25.08 (22.88–27.29) | 21.73 (20.72– 22.55) | 22.81(21.35–25.28) | <0.001 |
| Waist to hip circumference ratio | 0.88 ± 0.06 | 0.85 ± 0.05 | 0.88 ± 0.06 | <0.001 |
| HDL cholesterol, mmol/L | 1.31 (1.12–1.56) | 1.40 (1.31–1.49) | 1.40 (1.21–1.52) | 0.156 |
| LDL cholesterol, mmol/L | 2.8 (2.27–3.28) | 2.78 (2.50–3.12) | 2.79 (2.39–3.12) | 0.910 |
| TG, mmol/L | 1.4 (1.17–1.76) | 1.16 (0.90–1.79) | 1.29 (1.07–1.79) | 0.016 |
| TC, mmol/L | 4.87 ± 1.12 | 4.76 ± 0.57 | 4.82 ± 0.90 | 0.591 |
| Time to diagnosis | 1.44 (1.17–1.76) | NA | 1.44 (1.17–1.76) | |
| Histological subtypes | ||||
| Adenocarcinoma | 32 (78.05%) | NA | 32 (78.05%) | |
| Squamous cell carcinoma | 4 (9.76%) | NA | 4 (9.76%) | |
| Other subtypes | 5 (12.20%) | NA | 5 (12.20%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, Y.; Zhao, Q.; Wu, Y.; Liu, X.; Zhu, J.; Yu, Y.; Su, X.; Xu, K.; Jiang, Y.; Zhao, G. Serum Metabolomics Profiling Reveals Metabolic Alterations Prior to a Diagnosis with Non-Small Cell Lung Cancer among Chinese Community Residents: A Prospective Nested Case-Control Study. Metabolites 2022, 12, 906. https://doi.org/10.3390/metabo12100906
Xiang Y, Zhao Q, Wu Y, Liu X, Zhu J, Yu Y, Su X, Xu K, Jiang Y, Zhao G. Serum Metabolomics Profiling Reveals Metabolic Alterations Prior to a Diagnosis with Non-Small Cell Lung Cancer among Chinese Community Residents: A Prospective Nested Case-Control Study. Metabolites. 2022; 12(10):906. https://doi.org/10.3390/metabo12100906
Chicago/Turabian StyleXiang, Yu, Qi Zhao, Yilin Wu, Xing Liu, Junjie Zhu, Yuting Yu, Xuyan Su, Kelin Xu, Yonggen Jiang, and Genming Zhao. 2022. "Serum Metabolomics Profiling Reveals Metabolic Alterations Prior to a Diagnosis with Non-Small Cell Lung Cancer among Chinese Community Residents: A Prospective Nested Case-Control Study" Metabolites 12, no. 10: 906. https://doi.org/10.3390/metabo12100906