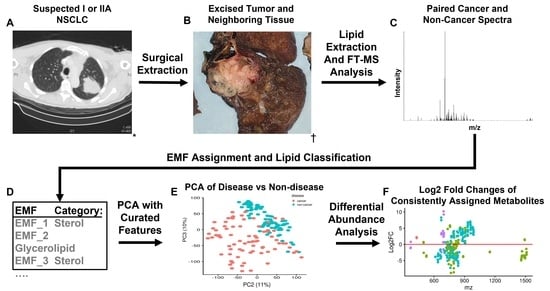
Untargeted Lipidomics of Non-Small Cell Lung Carcinoma Demonstrates Differentially Abundant Lipid Classes in Cancer vs. Non-Cancer Tissue
Abstract
:1. Introduction
2. Results
2.1. Mass Spectrometry Data Processing, Assignment Ambiguity and Quality Control of Samples
2.2. PCA and Sample Correlation Heatmap Shows Separation of Cancer and Non-Cancer Samples
2.3. Differential Abundance of Lipid Categories between Cancer and Non-Cancer Lung Tissue
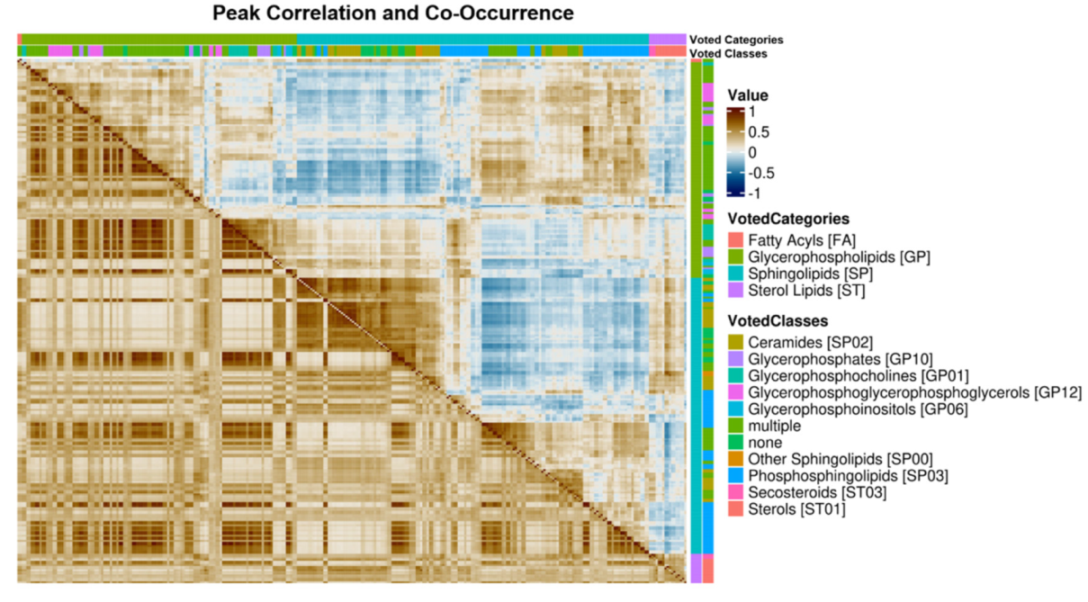
2.4. Lipid Category Correlation and Co-Occurrence Heatmaps
3. Discussion
3.1. Sample Correlation Analysis Shows Evidence of Metabolic Reprogramming in NSCLC
3.2. Regulatory Interpretation of Lipid Category Correlation and Co-Occurrence
3.3. Potential Clinical Implications
4. Materials and Methods
4.1. Description of Paired Human NSCLC Cancer Samples and Mass Spectrometry Analysis
4.2. Molecular Formula Assignment and Lipid Characterization of Assigned Formulas
4.3. Consistently Assigned Spectral Feature (Corresponded Peak) Generation and Peak Intensity Normalization
4.4. Quality Control of Patient Samples
4.5. Differential Abundance Analysis
4.6. Lipid Category Enrichment of Statistically Significant Peaks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Kanitkar, A.A.; Schwartz, A.G.; George, J.; Soubani, A.O. Causes of death in long-term survivors of non-small cell lung cancer: A regional Surveillance, Epidemiology, and End Results study. Ann. Thorac. Med. 2018, 13, 76. [Google Scholar]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo. Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Onoi, K.; Chihara, Y.; Uchino, J.; Shimamoto, T.; Morimoto, Y.; Iwasaku, M.; Kaneko, Y.; Yamada, T.; Takayama, K. Immune checkpoint inhibitors for lung cancer treatment: A review. J. Clin. Med. 2020, 9, 1362. [Google Scholar] [CrossRef]
- Gettinger, S.; Choi, J.; Hastings, K.; Truini, A.; Datar, I.; Sowell, R.; Wurtz, A.; Dong, W.; Cai, G.; Melnick, M.A. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 2017, 7, 1420–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.-j.; Du, Y.; Wen, R.; Yang, M.; Xu, J. Drug resistance to targeted therapeutic strategies in non-small cell lung cancer. Pharmacol. Ther. 2020, 206, 107438. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.J.; Soo, R.A. Resistance to immune checkpoint inhibitors in non-small cell lung cancer: Biomarkers and therapeutic strategies. Ther. Adv. Med. Oncol. 2020, 12, 1758835920937902. [Google Scholar] [CrossRef] [PubMed]
- SEER*Explorer: An interactive website for SEER cancer statistics. Available online: https://seer.cancer.gov/explorer/. (accessed on 15 March 2021).
- Uramoto, H.; Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242. [Google Scholar]
- Betticher, D.C. Adjuvant and neoadjuvant chemotherapy in NSCLC: A paradigm shift. Lung Cancer 2005, 50, S9–S16. [Google Scholar] [CrossRef]
- Pirker, R.; Krajnik, G.; Zöchbauer, S.; Malayeri, R.; Kneussl, M.; Huber, H. Paclitaxel/cisplatin in advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. 1995, 6, 833–835. [Google Scholar] [CrossRef]
- Sandler, A.B.; Nemunaitis, J.; Denham, C.; Von Pawel, J.; Cormier, Y.; Gatzemeier, U.; Mattson, K.; Manegold, C.; Palmer, M.; Gregor, A. Phase III trial of gemcitabine plus cisplatin versus cisplatin alone in patients with locally advanced or metastatic non–small-cell lung cancer. J. Clin. Oncol. 2000, 18, 122. [Google Scholar] [CrossRef]
- Vansteenkiste, J.; De Ruysscher, D.; Eberhardt, W.; Lim, E.; Senan, S.; Felip, E.; Peters, S.; Group, E.G.W. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013, 24, vi89–vi98. [Google Scholar] [CrossRef]
- Wozniak, A.J.; Crowley, J.J.; Balcerzak, S.P.; Weiss, G.R.; Spiridonidis, C.H.; Baker, L.H.; Albain, K.S.; Kelly, K.; Taylor, S.A.; Gandara, D.R. Randomized trial comparing cisplatin with cisplatin plus vinorelbine in the treatment of advanced non-small-cell lung cancer: A Southwest Oncology Group study. J. Clin. Oncol. 1998, 16, 2459–2465. [Google Scholar] [CrossRef]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, F.; Pereira, J.; Ciuleanu, T.; Tan, E.; Hirsh, V.; Thongprasert, S.; Bezjak, A.; Tu, D.; Santabarbara, P.; Seymour, L. A randomized placebo-controlled trial of erlotinib in patients with advanced non-small cell lung cancer (NSCLC) following failure of 1st line or 2nd line chemotherapy. A National Cancer Institute of Canada Clinical Trials Group (NCIC CTG) trial. J. Clin. Oncol. 2004, 22, 7022. [Google Scholar] [CrossRef]
- Chang, A.; Parikh, P.; Thongprasert, S.; Tan, E.H.; Perng, R.-P.; Ganzon, D.; Yang, C.-H.; Tsao, C.-J.; Watkins, C.; Botwood, N. Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non-small cell lung cancer: Subset analysis from the ISEL study. J. Thorac. Oncol. 2006, 1, 847–855. [Google Scholar] [CrossRef]
- Crino, L.; Kim, D.; Riely, G.; Janne, P.; Blackhall, F.; Camidge, D.; Hirsh, V.; Mok, T.; Solomon, B.; Park, K. Initial phase II results with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC): PROFILE 1005. J. Clin. Oncol. 2011, 29, 7514. [Google Scholar] [CrossRef]
- Ferrara, N.; Hillan, K.J.; Novotny, W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun. 2005, 333, 328–335. [Google Scholar] [CrossRef]
- Kim, D.-W.; Mehra, R.; Tan, D.S.-W.; Felip, E.; Chow, L.Q.M.; Camidge, D.R.; Vansteenkiste, J.F.; Sharma, S.; De Pas, T.; Riely, G.J.; et al. Ceritinib in advanced anaplastic lymphoma kinase (ALK)-rearranged (ALK+) non-small cell lung cancer (NSCLC): Results of the ASCEND-1 trial. J. Clin. Oncol. 2014, 32, 8003. [Google Scholar] [CrossRef]
- Piperdi, B.; Merla, A.; Perez-Soler, R. Targeting angiogenesis in squamous non-small cell lung cancer. Drugs 2014, 74, 403–413. [Google Scholar] [CrossRef] [Green Version]
- Sunshine, J.; Taube, J.M. Pd-1/Pd-L1 Inhibitors. Curr. Opin. Pharmacol. 2015, 23, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Sgambato, A.; Casaluce, F.; C Sacco, P.; Palazzolo, G.; Maione, P.; Rossi, A.; Ciardiello, F.; Gridelli, C. Anti PD-1 and PDL-1 immunotherapy in the treatment of advanced non-small cell lung cancer (NSCLC): A review on toxicity profile and its management. Curr. Drug Saf. 2016, 11, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Spigel, D.R.; Reckamp, K.L.; Rizvi, N.A.; Poddubskaya, E.; West, H.J.; Eberhardt, W.E.E.; Baas, P.; Antonia, S.J.; Pluzanski, A.; Vokes, E.E.; et al. A phase III study (CheckMate 017) of nivolumab (NIVO; anti-programmed death-1 [PD-1]) vs docetaxel (DOC) in previously treated advanced or metastatic squamous (SQ) cell non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2015, 33, 8009. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L. Metabolic heterogeneity in human lung tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellers, K.; Allen, T.D.; Bousamra, M.; Tan, J.; Méndez-Lucas, A.; Lin, W.; Bah, N.; Chernyavskaya, Y.; MacRae, J.I.; Higashi, R.M. Metabolic reprogramming and Notch activity distinguish between non-small cell lung cancer subtypes. Br. J. Cancer 2019, 121, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Sellers, K.; Fox, M.P.; Bousamra, M., II; Slone, S.P.; Higashi, R.M.; Miller, D.M.; Wang, Y.; Yan, J.; Yuneva, M.O.; Deshpande, R. Pyruvate carboxylase is critical for non–small-cell lung cancer proliferation. J. Clin. Investig. 2015, 125, 687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faubert, B.; Li, K.Y.; Cai, L.; Hensley, C.T.; Kim, J.; Zacharias, L.G.; Yang, C.; Do, Q.N.; Doucette, S.; Burguete, D. Lactate metabolism in human lung tumors. Cell 2017, 171, 358–371.e359. [Google Scholar] [CrossRef] [Green Version]
- Hassanein, M.; Hoeksema, M.D.; Shiota, M.; Qian, J.; Harris, B.K.; Chen, H.; Clark, J.E.; Alborn, W.E.; Eisenberg, R.; Massion, P.P. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin. Cancer Res. 2013, 19, 560–570. [Google Scholar] [CrossRef] [Green Version]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012, 481, 380–384. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, A.; Deng, X.; Khuri, F.R.; Owonikoko, T.K. Altered glutamine metabolism and therapeutic opportunities for lung cancer. Clin. Lung Cancer 2014, 15, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Osugi, J.; Yamaura, T.; Muto, S.; Okabe, N.; Matsumura, Y.; Hoshino, M.; Higuchi, M.; Suzuki, H.; Gotoh, M. Prognostic impact of the combination of glucose transporter 1 and ATP citrate lyase in node-negative patients with non-small lung cancer. Lung Cancer 2015, 88, 310–318. [Google Scholar] [CrossRef]
- Uramoto, H.; Osaki, T.; Inoue, M.; Taga, S.; Takenoyama, M.; Hanagiri, T.; Yoshino, I.; Nakanishi, R.; Ichiyoshi, Y.; Yasumoto, K. Fas expression in non-small cell lung cancer: Its prognostic effect in completely resected stage III patients. Eur. J. Cancer 1999, 35, 1462–1465. [Google Scholar] [CrossRef]
- Huang, J.; Fan, X.-X.; He, J.; Pan, H.; Li, R.-Z.; Huang, L.; Jiang, Z.; Yao, X.-J.; Liu, L.; Leung, E.L.-H. SCD1 is associated with tumor promotion, late stage and poor survival in lung adenocarcinoma. Oncotarget 2016, 7, 39970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csanadi, A.; Kayser, C.; Donauer, M.; Gumpp, V.; Aumann, K.; Rawluk, J.; Prasse, A.; zur Hausen, A.; Wiesemann, S.; Werner, M. Prognostic value of malic enzyme and ATP-citrate lyase in non-small cell lung cancer of the young and the elderly. PLoS ONE 2015, 10, e0126357. [Google Scholar] [CrossRef]
- Noto, A.; Raffa, S.; De Vitis, C.; Roscilli, G.; Malpicci, D.; Coluccia, P.; Di Napoli, A.; Ricci, A.; Giovagnoli, M.; Aurisicchio, L. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013, 4, e947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visca, P.; Sebastiani, V.; Botti, C.; Diodoro, M.G.; Lasagni, R.P.; Romagnoli, F.; Brenna, A.; De Joannon, B.C.; Donnorso, R.P.; Lombardi, G. Fatty acid synthase (FAS) is a marker of increased risk of recurrence in lung carcinoma. Anticancer Res. 2004, 24, 4169–4174. [Google Scholar]
- Wang, Y.; Zhang, X.; Tan, W.; Fu, J.; Zhang, W. Significance of fatty acid synthase expression in non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi Chin. J. Oncol. 2002, 24, 271–273. [Google Scholar]
- Sok, M.; Ravnik, J.; Ravnik, M. Preoperative total serum cholesterol as a prognostic factor for survival in patients with resectable non-small-cell lung cancer. Wien. Klin. Wochenschr. 2009, 121, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Jamnagerwalla, J.; Howard, L.E.; Allott, E.H.; Vidal, A.C.; Moreira, D.M.; Castro-Santamaria, R.; Andriole, G.L.; Freeman, M.R.; Freedland, S.J. Serum cholesterol and risk of high-grade prostate cancer: Results from the REDUCE study. Prostate Cancer Prostatic Dis. 2018, 21, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitahara, C.M.; de González, A.B.; Freedman, N.D.; Huxley, R.; Mok, Y.; Jee, S.H.; Samet, J.M. Total cholesterol and cancer risk in a large prospective study in Korea. J. Clin. Oncol. 2011, 29, 1592. [Google Scholar] [CrossRef] [Green Version]
- Ringerike, T.; Blystad, F.D.; Levy, F.O.; Madshus, I.H.; Stang, E. Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J. Cell Sci. 2002, 115, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.-S.; Chen, I.C.; Lee, C.-P.; Huang, R.-J.; Chen, P.-C.; Tsai, Y.-H.; Yang, Y.-H. Statin improves survival in patients with EGFR-TKI lung cancer: A nationwide population-based study. PLoS ONE 2017, 12, e0171137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.J.; Ezer, N.; Sigel, K.; Mhango, G.; Wisnivesky, J.P. The effect of statins on survival in patients with stage IV lung cancer. Lung Cancer 2016, 99, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.-Y.; Lee, S.-H.; Yoo, N.J.; Hyung, L.S.; Moon, Y.J.; Yun, T.; Kim, H.T.; Lee, J.S. A randomized phase II study of gefitinib plus simvastatin versus gefitinib alone in previously treated patients with advanced non–small cell lung cancer. Clin. Cancer Res. 2011, 17, 1553–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diomede, L.; Albani, D.; Sottocorno, M.; Donati, M.B.; Bianchi, M.; Fruscella, P.; Salmona, M. In vivo anti-inflammatory effect of statins is mediated by nonsterol mevalonate products. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1327–1332. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, M.M.; Tiglio, A.; Sadigh, K.; O’Donnell, L.; Collinge, M.; Pardi, R.; Bender, J.R. Inhibition of Interferon-Γ–Mediated Microvascular Endothelial Cell Major Histocompatibility Complex Class Ii Gene Activation by Hmg-Coa Reductase Inhibitors1. Transplantation 2001, 71, 1262–1268. [Google Scholar] [CrossRef]
- Weis, M.; Heeschen, C.; Glassford, A.J.; Cooke, J.P. Statins have biphasic effects on angiogenesis. Circulation 2002, 105, 739–745. [Google Scholar] [CrossRef] [Green Version]
- Tsoumpra, M.K.; Muniz, J.R.; Barnett, B.L.; Kwaasi, A.A.; Pilka, E.S.; Kavanagh, K.L.; Evdokimov, A.; Walter, R.L.; Von Delft, F.; Ebetino, F.H. The inhibition of human farnesyl pyrophosphate synthase by nitrogen-containing bisphosphonates. Elucidating the role of active site threonine 201 and tyrosine 204 residues using enzyme mutants. Bone 2015, 81, 478–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, J.M.; Flight, R.M.; Wang, Q.J.; Higashi, R.M.; Fan, T.W.-M.; Lane, A.N.; Moseley, H.N. New methods to identify high peak density artifacts in Fourier transform mass spectra and to mitigate their effects on high-throughput metabolomic data analysis. Metabolomics 2018, 14, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, J.M.; Flight, R.M.; Moseley, H.N. Small Molecule Isotope Resolved Formula Enumeration: A Methodology for Assigning Isotopologues and Metabolite Formulas in Fourier Transform Mass Spectra. Anal. Chem. 2019, 91, 8933. [Google Scholar] [CrossRef]
- Moseley, H.N.; Carreer, W.J.; Mitchell, J.; Flight, R.M. Method and system for identification of metabolites using mass spectra. U.S. Patent 10,607,723 B2, 31 March 2020. [Google Scholar]
- Mitchell, J.M.; Fan, T.W.M.; Lane, A.N.; Moseley, H.N.B. Development and in silico evaluation of large-scale metabolite identification methods using functional group detection for metabolomics. Front. Genet. 2014, 5, 237. [Google Scholar] [CrossRef] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, J.M.; Flight, R.M.; Moseley, H.N. Deriving Lipid Classification Based on Molecular Formulas. Metabolites 2020, 10, 122. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, C.; Oh, D.S.; Marron, J.; He, X.; Qaqish, B.F.; Livasy, C.; Carey, L.A.; Reynolds, E.; Dressler, L. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006, 7, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011, 43, 768. [Google Scholar] [CrossRef] [Green Version]
- Mizumachi, T.; Muskhelishvili, L.; Naito, A.; Furusawa, J.; Fan, C.Y.; Siegel, E.R.; Kadlubar, F.F.; Kumar, U.; Higuchi, M. Increased distributional variance of mitochondrial DNA content associated with prostate cancer cells as compared with normal prostate cells. Prostate 2008, 68, 408–417. [Google Scholar] [CrossRef] [Green Version]
- Ericsson, J.; Jackson, S.M.; Kim, J.B.; Spiegelman, B.M.; Edwards, P.A. Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1-and sterol regulatory element-binding protein-responsive gene. J. Biol. Chem. 1997, 272, 7298–7305. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.-A.; Xiong, X.; Zaytseva, Y.Y.; Napier, D.L.; Vallee, E.; Li, A.T.; Wang, C.; Weiss, H.L.; Evers, B.M.; Gao, T. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018, 9, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, D.; Xiao, H.; Dong, J.; Li, Y.; Feng, G.; Cui, M.; Fan, S. B7-H3 regulates lipid metabolism of lung cancer through SREBP1-mediated expression of FASN. Biochem. Biophys. Res. Commun. 2017, 482, 1246–1251. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Royaux, I.; Swinnen, J.V.; Smans, K. ATP citrate lyase knockdown induces growth arrest and apoptosis through different cell-and environment-dependent mechanisms. Mol. Cancer Ther. 2012, 11, 1925–1935. [Google Scholar] [CrossRef] [Green Version]
- Gabitova, L.; Gorin, A.; Astsaturov, I. Molecular pathways: Sterols and receptor signaling in cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 20, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Sukhanova, A.; Gorin, A.; Serebriiskii, I.G.; Gabitova, L.; Zheng, H.; Restifo, D.; Egleston, B.L.; Cunningham, D.; Bagnyukova, T.; Liu, H. Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov. 2013, 3, 96–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bené, H.; Lasky, D.; Ntambi, J.M. Cloning and characterization of the human stearoyl-CoA desaturase gene promoter: Transcriptional activation by sterol regulatory element binding protein and repression by polyunsaturated fatty acids and cholesterol. Biochem. Biophys. Res. Commun. 2001, 284, 1194–1198. [Google Scholar] [CrossRef]
- Li, J.; Gu, D.; Lee, S.S.; Song, B.; Bandyopadhyay, S.; Chen, S.; Konieczny, S.F.; Ratliff, T.L.; Liu, X.; Xie, J. Abrogating cholesterol esterification suppresses growth and metastasis of pancreatic cancer. Oncogene 2016, 35, 6378–6388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.M.; Zhao, Y.; Zhang, J.P.; Zhang, J.H.; Jiang, P.; Li, B.; Wang, C. Expression and potential mechanism of metabolism-related genes and CRLS1 in non-small cell lung cancer. Oncol. Lett. 2018, 15, 2661–2668. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.S.; Kotha, S.R.; Avasarala, S.; VanScoyk, M.; Winn, R.A.; Pennathur, A.; Yashaswini, P.S.; Bandela, M.; Salgia, R.; Tyurina, Y.Y. Lysocardiolipin acyltransferase regulates NSCLC cell proliferation and migration by modulating mitochondrial dynamics. J. Biol. Chem. 2020, 295, 13393–13406. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.H.-R.; Yang, C.; Tse, A.P.-W.; Wei, L.; Lee, D.; Zhang, M.S.; Goh, C.C.; Chiu, D.K.-C.; Yuen, V.W.-H.; Law, C.-T. Genome-wide CRISPR-Cas9 knockout library screening identified PTPMT1 in cardiolipin synthesis is crucial to survival in hypoxia in liver cancer. Cell Rep. 2021, 34, 108676. [Google Scholar] [CrossRef]
- Sheng, J.; Zhao, Q.; Zhao, J.; Zhang, W.; Sun, Y.; Qin, P.; Lv, Y.; Bai, L.; Yang, Q.; Chen, L. SRSF1 modulates PTPMT1 alternative splicing to regulate lung cancer cell radioresistance. EBioMedicine 2018, 38, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Dudek, J. Role of cardiolipin in mitochondrial signaling pathways. Front. Cell Dev. Biol. 2017, 5, 90. [Google Scholar] [CrossRef] [Green Version]
- Ponnusamy, S.; Meyers-Needham, M.; Senkal, C.E.; Saddoughi, S.A.; Sentelle, D.; Selvam, S.P.; Salas, A.; Ogretmen, B. Sphingolipids and cancer: Ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol. 2010, 6, 1603–1624. [Google Scholar] [CrossRef] [Green Version]
- Flight, R.M.; Moseley, H.N.B. ICIKendallTau. Available online: https://github.com/MoseleyBioinformaticsLab/ICIKendallTau (accessed on 23 July 2021).
- Gierliński, M.; Cole, C.; Schofield, P.; Schurch, N.J.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.; Owen-Hughes, T. Statistical models for RNA-seq data derived from a two-condition 48-replicate experiment. Bioinformatics 2015, 31, 3625–3630. [Google Scholar] [CrossRef]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Li, Y.; Fan, T.W.; Lane, A.N.; Kang, W.-Y.; Arnold, S.M.; Stromberg, A.J.; Wang, C.; Chen, L. SDA: A semi-parametric differential abundance analysis method for metabolomics and proteomics data. BMC Bioinform. 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, C.; Chen, L. SDAMS: Differential Abundant Analysis for Metabolomics, Proteomics and Single-Cell RNA Sequencing Data., R Package Version 1.10.0; 2020. Available online: https://bioconductor.org/packages/devel/bioc/manuals/SDAMS/man/SDAMS.pdf (accessed on 15 March 2015).
- Thissen, D.; Steinberg, L.; Kuang, D. Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J. Educ. Behav. Stat. 2002, 27, 77–83. [Google Scholar] [CrossRef]
- Sakai, R.; Winand, R.; Verbeiren, T.; Moere, A.V.; Aerts, J. dendsort: Modular leaf ordering methods for dendrogram representations in R. F1000Research 2014, 3, 177. [Google Scholar] [CrossRef] [Green Version]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Flight, R.M.; Harrison, B.J.; Mohammad, F.; Bunge, M.B.; Moon, L.D.; Petruska, J.C.; Rouchka, E.C. categoryCompare, an analytical tool based on feature annotations. Front. Genet. 2014, 5, 98. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.M. Computational Tools for the Untargeted Assignment of FT-MS Computational Tools for the Untargeted Assignment of FT-MS Metabolomics Datasets. Ph.D. Thesis, University of Kentucky, Lexington, KY, USA, 2019; p. 42. [Google Scholar]




| Category | Total | More Abundant Features | Less Abundant Features | ||||
|---|---|---|---|---|---|---|---|
| Expected | Observed | p-Adjust | Expected | Observed | p-Adjust | ||
| Fatty Acyls [FA] | 12 | 2.989 | 2 | 1 | 3.947 | 0 | 1 |
| Glycerophospholipids [GP] | 205 | 51.055 | 37 | 1 | 67.424 | 88 | 0.00503 |
| Prenol Lipids [PR] | 5 | 1.245 | 0 | 1 | 1.644 | 0 | 1 |
| Sphingolipids [SP] | 281 | 69.983 | 79 | 0.09861 | 92.420 | 81 | 1 |
| Sphingolipids [SP]–Low m/z | 33 | 8.219 | 3 | 1 | 10.854 | 16 | 0.141 |
| Sphingolipids [SP]–High m/z | 248 | 61.764 | 76 | 0.00967 | 81.567 | 65 | 1 |
| Sterol Lipids [ST] | 23 | 5.728 | 13 | 0.00643 | 7.084 | 3 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell, J.M.; Flight, R.M.; Moseley, H.N.B. Untargeted Lipidomics of Non-Small Cell Lung Carcinoma Demonstrates Differentially Abundant Lipid Classes in Cancer vs. Non-Cancer Tissue. Metabolites 2021, 11, 740. https://doi.org/10.3390/metabo11110740
Mitchell JM, Flight RM, Moseley HNB. Untargeted Lipidomics of Non-Small Cell Lung Carcinoma Demonstrates Differentially Abundant Lipid Classes in Cancer vs. Non-Cancer Tissue. Metabolites. 2021; 11(11):740. https://doi.org/10.3390/metabo11110740
Chicago/Turabian StyleMitchell, Joshua M., Robert M. Flight, and Hunter N. B. Moseley. 2021. "Untargeted Lipidomics of Non-Small Cell Lung Carcinoma Demonstrates Differentially Abundant Lipid Classes in Cancer vs. Non-Cancer Tissue" Metabolites 11, no. 11: 740. https://doi.org/10.3390/metabo11110740