Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective
Abstract
:1. Introduction: Problem of Soil Salinization and Impact of Salinity on Plants
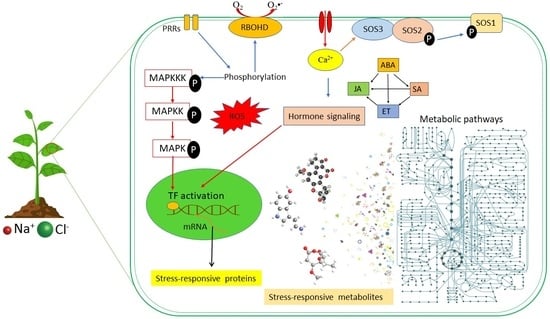
2. Plant Responses to Salinity Stress: Cellular and Molecular Events
2.1. Signalling Pathways in Salinity Stress Conditions
2.1.1. Second Messenger Signalling: ROS and Ca2+ Waves
2.1.2. The SOS Signalling Pathway
2.1.3. Hormonal Crosstalk in Salinity Responses
3. Metabolomics for Elucidation of Plant Responses to Salinity
Plant Responses to Salinity: Metabolic Reprogramming
4. Conclusions and Prospective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zaman, M.; Shahid, S.A.; Pharis, R.P. Salinity a Serious Threat to Food Security—Where Do We Stand? Int. At. Energy Agency Bull. 2016, 39, 9–10. [Google Scholar]
- García-Mauriño, S.; Monreal, J.A.; Alvarez, R.; Vidal, J.; Echevarría, C. Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: Independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 2003, 216, 648–655. [Google Scholar] [CrossRef]
- Kissoudis, C.; Van De Wiel, C.; Visser, R.G.F.; Van Der Linden, G. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant. Sci. 2014, 5, 207. [Google Scholar] [CrossRef] [Green Version]
- Shokat, S.; Großkinsky, D.K. Tackling Salinity in Sustainable Agriculture—What Developing Countries May Learn from Approaches of the Developed World the saline Global Area under salinity (Million Hectares). Sustainability 2019, 11, 4558. [Google Scholar] [CrossRef] [Green Version]
- Wichelns, D.; Qadir, M. Achieving sustainable irrigation requires effective management of salts, soil salinity, and shallow groundwater. Agric. Water Manag. 2015, 157, 31–38. [Google Scholar] [CrossRef]
- Kumar, P.; Sharma, P.K. Soil Salinity and Food Security in India. Front. Sustain. Food Syst. 2020, 4, 533781. [Google Scholar] [CrossRef]
- Aras, S.; Keles, H.; Eşitken, A. Silicon nutrition counteracts salt-induced damage associated with changes in biochemical responses in apple. Bragantia 2020, 79, 1–7. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Higgs, D.; Murillo-Amador, B.; Aydemir, S.; Girgin, A.R. Silicon improves salinity tolerance in wheat plants. Environ. Exp. Bot. 2008, 62, 10–16. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Abdelraheem, A.; Esmaeili, N.; O’Connell, M.; Zhang, J. Progress and perspective on drought and salt stress tolerance in cotton. Ind. Crops Prod. 2019, 130, 118–129. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Ghassemi-Golezani, K. Improving amino acid composition of soybean under salt stress by salicylic acid and jasmonic acid. J. Appl. Bot. Food Qual. 2016, 89, 243–248. [Google Scholar] [CrossRef]
- Bistgani, Z.E.; Hashemi, M.; DaCosta, M.; Craker, L.; Maggi, F.; Morshedloo, M.R. Effect of salinity stress on the physiological characteristics, phenolic compounds and antioxidant activity of Thymus vulgaris L. and Thymus daenensis Celak. Ind. Crops Prod. 2019, 135, 311–320. [Google Scholar] [CrossRef]
- Savvas, D.; Ntatsi, G. Biostimulant activity of silicon in horticulture. Sci. Hortic. 2015, 196, 66–81. [Google Scholar] [CrossRef]
- Afzal, M.Z.; Jia, Q.; Ibrahim, A.K.; Niyitanga, S.; Zhang, L. Mechanisms and Signaling Pathways of Salt Tolerance in Crops: Understanding from the Transgenic Plants. Trop. Plant Biol. 2020, 13, 297–320. [Google Scholar] [CrossRef]
- Basu, S.; Rabara, R. Abscisic acid—An enigma in the abiotic stress tolerance of crop plants. Plant Gene 2017, 11, 90–98. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torabi, F.; Majd, A.; Enteshari, S.; Torabi, F.; Majd, A.; Enteshari, S. Soil Science and Plant Nutrition The effect of silicon on alleviation of salt stress in borage (Borago officinalis L.) The effect of silicon on alleviation of salt stress in borage (Borago officinalis L.). Soil Sci. Plant Nutr. 2015, 61, 788–798. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Soundararajan, P.; Manivannan, A. Mechanisms of silicon-mediated amelioration of salt stress in plants. Plants 2019, 8, 307. [Google Scholar] [CrossRef] [Green Version]
- Evelin, H.; Devi, T.S.; Gupta, S.; Kapoor, R. Mitigation of Salinity Stress in Plants by Arbuscular Mycorrhizal Symbiosis: Current Understanding and New Challenges. Front. Plant Sci. 2019, 10, 470. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, G.; Fan, X.; Peng, M.; Yin, C.; Xiao, Z.; Liang, Y. Silicon Improves Rice Salinity Resistance by Alleviating Ionic Toxicity and Osmotic Constraint in an Organ-Specific Pattern. Front. Plant Sci. 2020, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Salt, D.E.E. Plant Ionomics: From Elemental Profiling to Environmental Adaptation. Mol. Plant 2016, 9, 787–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sévin, D.C.; Kuehne, A.; Zamboni, N.; Sauer, U. Biological insights through nontargeted metabolomics. Curr. Opin. Biotechnol. 2015, 34, 1–8. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A conversation on data mining strategies in LC-MS untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.-P.; Chao, D.-Y.; Lin, H.-X. Toward Understanding Molecular Mechanisms of Abiotic Stress Responses in Rice. Rice 2008, 1, 36–51. [Google Scholar] [CrossRef] [Green Version]
- Demidchik, V. ROS-activated ion channels in plants: Biophysical characteristics, physiological functions and molecular nature. Int. J. Mol. Sci. 2018, 19, 1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Saijo, Y.; Loo, E.P. iian Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhise, K.K.; Dandge, P.B. Mitigation of salinity stress in plants using plant growth promoting bacteria. Symbiosis 2019, 79, 191–204. [Google Scholar] [CrossRef]
- Zhu, J. Review Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.-P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [Green Version]
- Nejat, N.; Mantri, N. Plant Immune System: Crosstalk Between Responses to Biotic and Abiotic Stresses the Missing Link in Understanding Plant Defence. Curr. Issues Mol. Biol. 2017, 23, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, S.; Kaya, H.; Kawarazaki, T.; Hiraoka, G.; Senzaki, E.; Michikawa, M.; Kuchitsu, K. Protein phosphorylation is a prerequisite for the Ca 2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca 2+ and reactive oxygen species. Biochim. Biophys. Acta—Mol. Cell Res. 2012, 1823, 398–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, N.; Miller, G.; Salazar, C.; Mondal, H.A.; Shulaev, E.; Cortes, D.F.; Shuman, J.L.; Luo, X.; Shah, J.; Schlauch, K.; et al. Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 2013, 25, 3553–3569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.G.; Miller, G.; Wallace, I.; Harper, J.; Mittler, R.; Gilroy, S. Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J. 2017, 90, 698–707. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 2018, 154, 134–142. [Google Scholar] [CrossRef]
- Kibria, M.G.; Hossain, M.; Murata, Y.; Hoque, M.A. Antioxidant Defense Mechanisms of Salinity Tolerance in Rice Genotypes. Rice Sci. 2017, 24, 155–162. [Google Scholar] [CrossRef]
- Ahmad, P.; Prasad, M.N.V. Environmental Adaptations and Stress Tolerance of Plants in the Era of Climate Change; Ahmad, P., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2012; ISBN 978-1-4614-0814-7. [Google Scholar]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Fountain, J.C.; Ji, P.; Ni, X.; Chen, S.; Lee, R.D.; Kemerait, R.C.; Guo, B. Deciphering drought-induced metabolic responses and regulation in developing maize kernels. Plant Biotechnol. J. 2018, 16, 1616–1628. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, D.; Zhou, R.; Wang, X.; Dossa, K.; Wang, L.; Zhang, Y.; Yu, J.; Gong, H.; Zhang, X.; et al. Transcriptome and metabolome analyses of two contrasting sesame genotypes reveal the crucial biological pathways involved in rapid adaptive response to salt stress. BMC Plant Biol. 2019, 19, 66. [Google Scholar] [CrossRef] [Green Version]
- Gimenez, E.; Salinas, M.; Manzano-Agugliaro, F. Worldwide research on plant defense against biotic stresses as improvement for sustainable agriculture. Sustainability 2018, 10, 391. [Google Scholar] [CrossRef] [Green Version]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef] [Green Version]
- Mühlenbock, P.; Szechyńska-Hebda, M.; Płaszczyca, M.; Baudo, M.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpińskie, S. Chloroplast signaling and lesion simulating disease1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef] [Green Version]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant, Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca 2+ and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef]
- Tuteja, N.; Mahajan, S. Calcium signaling network in plants: An overview. Plant Signal. Behav. 2007, 2, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, F.; Shinozaki, K. Long-distance signaling in plant stress response. Curr. Opin. Plant Biol. 2019, 47, 106–111. [Google Scholar] [CrossRef]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [CrossRef]
- Dodd, A.N.; Kudla, J.; Sanders, D. The Language of Calcium Signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.-K. Molecular Characterization of Functional Domains in the Protein Kinase SOS2 That Is Required for Plant Salt Tolerance. Plant Cell 2007, 13, 1383. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca 2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef] [PubMed]
- Drerup, M.M.; Schlücking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpinska, B.; Zhang, K.; Rasool, B.; Pastok, D.; Morris, J.; Verrall, S.R.; Hedley, P.E.; Hancock, R.D.; Foyer, C.H. The redox state of the apoplast influences the acclimation of photosynthesis and leaf metabolism to changing irradiance. Plant Cell Environ. 2018, 41, 1083–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, calcium, and electric signals: Key mediators of rapid systemic signaling in plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Dong, C.H.; Zolman, B.K.; Bartel, B.; Lee, B.H.; Stevenson, B.; Agarwal, M.; Zhu, J.K. Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol. Plant 2009, 2, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Knight, H.; Knight, M.R.; Knight, H. Abiotic stress signalling pathways: Specificity and cross-talk. Trends. Plant. Sci. 2001, 6, 262–267. [Google Scholar] [CrossRef]
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP Kinase Cascades in Arabidopsis Innate Immunity. Front. Plant Sci. 2012, 3, 169. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef]
- Mahajan, S.; Pandey, G.K.; Tuteja, N. Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 2008, 471, 146–158. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaki, F.; Maboud, H.E.; Niknam, V. Effects of salicylic acid on hormonal cross talk, fatty acids profile, and ions homeostasis from salt-stressed safflower. J. Plant Interact. 2019, 14, 340–346. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P.; Hasanuzzaman, M.; Fujita, M. Molecular Sciences Regulation of Plant Responses to Salt Stress. J. Mol. Sci 2021, 22, 4609. [Google Scholar] [CrossRef]
- Dange, M.C.; Mishra, V.; Mukherjee, B.; Jaiswal, D.; Merchant, M.S.; Prasannan, C.B.; Wangikar, P.P. Evaluation of freely available software tools for untargeted quantification of 13C isotopic enrichment in cellular metabolome from HR-LC/MS data. Metab. Eng. Commun. 2020, 10, e00120. [Google Scholar] [CrossRef]
- He, J.; Duan, Y.; Hua, D.; Fan, G.; Wang, L.; Liu, Y.; Chen, Z.; Han, L.; Qu, L.-J.; Gong, Z. DEXH Box RNA Helicase-Mediated Mitochondrial Reactive Oxygen Species Production in Arabidopsis Mediates Crosstalk between Abscisic Acid and Auxin Signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef] [Green Version]
- Dresselhaus, T.; Hückelhoven, R. Biotic and Abiotic Stress Responses in Crop Plants. Agronomy 2018, 8, 267. [Google Scholar] [CrossRef] [Green Version]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd-Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef] [Green Version]
- Kohli, A.; Sreenivasulu, N.; Lakshmanan, P.; Kumar, P.P. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 2013, 32, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Maksymiec, W. Signaling responses in plants to heavy metal stress. Acta Physiol. Plant. 2007, 29, 177–187. [Google Scholar] [CrossRef]
- Larkindale, J.; Huang, B. Thermotolerance and antioxidant systems in Agrostis stolonifera: Involvement of salicylic acid, abscisic acid, calcium, hydrogen peroxide, and ethylene. J. Plant Physiol. 2004, 161, 405–413. [Google Scholar] [CrossRef]
- Adiloğlu, S. We are IntechOpen, the world ’ s leading publisher of Open Access books Built by scientists, for scientists TOP 1%. Heavy Met. Remov. Phytoremediat. 2016, i, 13. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, S.; Shah, S.; Kumar, R.; Vashisth, D.; Akhtar, M.Q.; Kumar, A.; Dwivedi, U.N.; Shasany, A.K. Ocimum metabolomics in response to abiotic stresses: Cold, flood, drought and salinity. PLoS ONE 2019, 14, 1–26. [Google Scholar] [CrossRef]
- Munemasa, S.; Oda, K.; Watanabe-Sugimoto, M.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 2007, 143, 1398–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, G.K. Emergence of a novel calcium signaling pathway in plants: CBL-CIPK signaling network. Physiol. Mol. Biol. Plants 2008, 14, 51–68. [Google Scholar] [CrossRef] [Green Version]
- Asgher, M.; Khan, M.I.R.; Anjum, N.A.; Khan, N.A. Minimising toxicity of cadmium in plants—role of plant growth regulators. Protoplasma 2015, 252, 399–413. [Google Scholar] [CrossRef]
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.H.; Kim, Y.S.; Park, S.H.; Koo, Y.J.; Choi, Y.D.; Chung, Y.Y.; Lee, I.J.; Kim, J.K. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice 1[W][OA]. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef] [Green Version]
- Elsheery, N.I.; Helaly, M.N.; Omar, S.A.; John, S.V.S.; Zabochnicka-Swiątek, M.; Kalaji, H.M.; Rastogi, A. Physiological and molecular mechanisms of salinity tolerance in grafted cucumber. S. Afr. J. Bot. 2020, 130, 90–102. [Google Scholar] [CrossRef]
- Manaa, A.; Gharbi, E.; Mimouni, H.; Wasti, S.; Aschi-Smiti, S.; Lutts, S.; Ben Ahmed, H. Simultaneous application of salicylic acid and calcium improves salt tolerance in two contrasting tomato (Solanum lycopersicum) cultivars. S. Afr. J. Bot. 2014, 95, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Zhang, C.; Chu, L.Y.; Shao, H.B. Responses of higher plants to abiotic stresses and agricultural sustainable development. J. Plant Interact. 2007, 2, 135–147. [Google Scholar] [CrossRef]
- Iqbal, M.; Ashraf, M. Gibberellic acid mediated induction of salt tolerance in wheat plants: Growth, ionic partitioning, photosynthesis, yield and hormonal homeostasis. Environ. Exp. Bot. 2013, 86, 76–85. [Google Scholar] [CrossRef]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar] [CrossRef]
- Venegas-Molina, J.; Proietti, S.; Pollier, J.; Orozco-Freire, W.; Ramirez-Villacis, D.; Leon-Reyes, A. Induced tolerance to abiotic and biotic stresses of broccoli and Arabidopsis after treatment with elicitor molecules. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, J.; Zhu, T.; Zhao, C.; Li, L.; Chen, M. The role of melatonin in salt stress responses. Int. J. Mol. Sci. 2019, 20, 1735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munné-Bosch, S.; Müller, M. Hormonal cross-talk in plant development and stress responses. Front. Plant Sci. 2013, 4, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugizimana, F.; Engel, J.; Salek, R.; Dubery, I.; Piater, L.; Burgess, K. The Disruptive 4IR in the Life Sciences: Metabolomics; Springer International Publishing: Cham, Switzerland, 2020; Volume 674, ISBN 9783030482305. [Google Scholar]
- Banerjee, A.; Roychoudhury, A. Epigenetic regulation during salinity and drought stress in plants: Histone modifications and DNA methylation. Plant Gene 2017, 11, 199–204. [Google Scholar] [CrossRef]
- Quan, X.; Liu, J.; Zhang, N.; Xie, C.; Li, H.; Xia, X.; He, W.; Qin, Y. Genome-Wide Association Study Uncover the Genetic Architecture of Salt Tolerance-Related Traits in Common Wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 663941. [Google Scholar] [CrossRef]
- Chen, J.T.; Aroca, R.; Romano, D. Molecular aspects of plant salinity stress and tolerance. Int. J. Mol. Sci. 2021, 22, 4918. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; Rangani, J.; Parida, A.K. Unraveling salt responsive metabolites and metabolic pathways using non-targeted metabolomics approach and elucidation of salt tolerance mechanisms in the xero-halophyte Haloxylon salicornicum. Plant Physiol. Biochem. 2021, 158, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Khan, M.S.; Davies, E. PlantOmics: The Omics of Plant Science; Barh, D., Khan, M.S., Davies, E., Eds.; Springer: New Delhi, India, 2015; ISBN 978-81-322-2171-5. [Google Scholar]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [Green Version]
- Low, C.F.; Rozaini, M.Z.H.; Musa, N.; Syarul Nataqain, B. Current knowledge of metabolomic approach in infectious fish disease studies. J. Fish Dis. 2017, 40, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, W.; Chen, Q.; Ding, R. Effects of silicon on H +-ATPase and H +-PPase activity, fatty acid composition and fluidity of tonoplast vesicles from roots of salt-stressed barley (Hordeum vulgare L.). Environ. Exp. Bot. 2005, 53, 29–37. [Google Scholar] [CrossRef]
- Majumder, E.L.W.; Billings, E.M.; Benton, H.P.; Martin, R.L.; Palermo, A.; Guijas, C.; Rinschen, M.M.; Domingo-Almenara, X.; Montenegro-Burke, J.R.; Tagtow, B.A.; et al. Cognitive analysis of metabolomics data for systems biology. Nat. Protoc. 2021, 16, 1376–1418. [Google Scholar] [CrossRef]
- Ullah, N.; Yüce, M.; Neslihan Öztürk Gökçe, Z.; Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genomics 2017, 18, 969. [Google Scholar] [CrossRef]
- Madsen, R.; Lundstedt, T.; Trygg, J. Chemometrics in metabolomics-A review in human disease diagnosis. Anal. Chim. Acta 2010, 659, 23–33. [Google Scholar] [CrossRef]
- Pandey, G.K.; Pandey, A.; Prasad, M.; Böhmer, M. Editorial: Abiotic Stress Signaling in Plants: Functional Genomic Intervention. Front. Plant Sci. 2016, 7, 681. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Alakwaa, F.M.; Chaudhary, K.; Garmire, L.X. Deep Learning Accurately Predicts Estrogen Receptor Status in Breast Cancer Metabolomics Data. J. Proteome Res. 2018, 17, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol. Plant. 2008, 132, 117–135. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Kumar, N.; Mukhtar, M.S. Systems biology and machine learning in plant–pathogen interactions. Mol. Plant-Microbe Interact. 2019, 32, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Ying, H.; Shi, J.; Zhang, S.; Pingcuo, G.; Wang, S.; Zhao, F.; Cui, Y.; Zeng, X. Transcriptomic and metabolomic profiling provide novel insights into fruit development and flesh coloration in Prunus mira Koehne, a special wild peach species. BMC Plant Biol. 2019, 19, 463. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant. Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [Green Version]
- Rai, A.; Yamazaki, M.; Saito, K. A new era in plant functional genomics. Curr. Opin. Syst. Biol. 2019, 15, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Quanbeck, S.M.; Brachova, L.; Campbell, A.A.; Guan, X.; Perera, A.; He, K.; Rhee, S.Y.; Bais, P.; Dickerson, J.A.; Dixon, P.; et al. Metabolomics as a hypothesis-generating functional genomics tool for the annotation of Arabidopsis thaliana genes of “unknown function”. Front. Plant Sci. 2012, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant–Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Schwachtje, J.; Fischer, A.; Erban, A.; Kopka, J. Primed primary metabolism in systemic leaves: A functional systems analysis. Sci. Rep. 2018, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta—Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef] [Green Version]
- Brunetti, C.; George, R.M.; Tattini, M.; Field, K.; Davey, M.P. Metabolomics in plant environmental physiology. J. Exp. Bot. 2013, 64, 4011–4020. [Google Scholar] [CrossRef] [Green Version]
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 29265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Casati, D.F.; Zanor, M.I.; Busi, M.V. Metabolomics in plants and humans: Applications in the prevention and diagnosis of diseases. Biomed Res. Int. 2013, 2013, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyne, M.E.; Narcross, L.; Martin, V.J.J. Engineering Plant Secondary Metabolism in Microbial Systems. Plant Physiol. 2019, 179, 844–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boccard, J.; Rudaz, S. Harnessing the complexity of metabolomic data with chemometrics. J. Chemom. 2014, 28, 1–9. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. TrAC Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Pan, J.; Li, Z.; Dai, S.; Ding, H.; Wang, Q.; Li, X.; Ding, G.; Wang, P.; Guan, Y.; Liu, W. Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep. 2020, 10, 13660. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Gong, H.J.; Yin, J.L. Role of silicon in mediating salt tolerance in plants: A Review. Plants 2019, 8, 147. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for plant stress response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Kachlicki, P.; Stobiecki, M. Analytical methods for detection of plant metabolomes changes in response to biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razzaq, A.; Sadia, B.; Raza, A.; Hameed, M.K.; Saleem, F. Metabolomics: A way forward for crop improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.; King, R.D.; Altmann, T.; Fiehn, O. Discrimination Using Statistics and Machine Learning. Bioinformatics 2002, 18, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Król, A.; Amarowicz, R.; Weidner, S. Changes in the composition of phenolic compounds and antioxidant properties of grapevine roots and leaves (vitis viniferal.) under continuous of long-term drought stress. Acta Physiol. Plant. 2014, 36, 1491–1499. [Google Scholar] [CrossRef] [Green Version]
- Silva, E.; da Graça, J.P.; Porto, C.; Martin do Prado, R.; Hoffmann-Campo, C.B.; Meyer, M.C.; de Oliveira Nunes, E.; Pilau, E.J. Unraveling Asian Soybean Rust metabolomics using mass spectrometry and Molecular Networking approach. Sci. Rep. 2020, 10, 138. [Google Scholar] [CrossRef]
- Wu, D.; Cai, S.; Chen, M.; Ye, L.; Chen, Z.; Zhang, H.; Dai, F.; Wu, F.; Zhang, G. Tissue Metabolic Responses to Salt Stress in Wild and Cultivated Barley. PLoS ONE 2013, 8, e055431. [Google Scholar] [CrossRef] [PubMed]
- Bandehagh, A.; Taylor, N.L. Can Alternative Metabolic Pathways and Shunts Overcome Salinity Induced Inhibition of Central Carbon Metabolism in Crops? Front. Plant Sci. 2020, 11, 1072. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Huan, L.; Gao, S.; He, L.; Wang, G. NADPH from the oxidative pentose phosphate pathway drives the operation of cyclic electron flow around photosystem I in high-intertidal macroalgae under severe salt stress. Physiol. Plant. 2016, 156, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Huan, L.; Xie, X.; Zheng, Z.; Sun, F.; Wu, S.; Li, M.; Gao, S.; Gu, W.; Wang, G. Positive correlation between psi response and oxidative pentose phosphate pathway activity during salt stress in an intertidal macroalga. Plant Cell Physiol. 2014, 55, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Manna, I.; Sil, P.; Bandyopadhyay, M.; Biswas, A.K. Exogenous silicon alters organic acid production and enzymatic activity of TCA cycle in two NaCl stressed indica rice cultivars. Plant Physiol. Biochem. 2019, 136, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, T.M. Synthesis versus degradation: Directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol. Biol. 2018, 98, 121–135. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, Y.; Wang, X.; Liu, J. Novel insights into salinity-induced lipogenesis and carotenogenesis in the oleaginous astaxanthin-producing alga Chromochloris zofingiensis: A multi-omics study. Biotechnol. Biofuels 2020, 13, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.C.; Park, J.C.; Kim, D.H.; Kang, S.; Shin, K.H.; Park, H.G.; Han, J.; Lee, J.S. Interrelationship of salinity shift with oxidative stress and lipid metabolism in the monogonont rotifer Brachionus koreanus. Comp. Biochem. Physiol.—Part A Mol. Integr. Physiol. 2017, 214, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Candeias, N.R.; Assoah, B.; Simeonov, S.P. Production and Synthetic Modifications of Shikimic Acid. Chem. Rev. 2018, 118, 10458–10550. [Google Scholar] [CrossRef] [PubMed]
- Naikoo, M.I.; Dar, M.I.; Raghib, F.; Jaleel, H.; Ahmad, B.; Raina, A.; Khan, F.A.; Naushin, F. Role and regulation of plants phenolics in abiotic stress tolerance: An overview. Plant Signal. Mol. Role Regul. Stress. Environ. 2019, 157–168. [Google Scholar] [CrossRef]
- Xu, N.; Liu, S.; Lu, Z.; Pang, S.; Wang, L.; Wang, L.; Li, W. Gene expression profiles and flavonoid accumulation during salt stress in ginkgo biloba seedlings. Plants 2020, 9, 1162. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X.; Li, F.; Wang, Y.; Wu, Z.; Fu, C.; Lin, H.; et al. NTMYB4 and NTCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Nada, K.; Honda, C.; Kitashiba, H.; Wen, X.P.; Pang, X.M.; Moriguchi, T. Polyamine biosynthesis of apple callus under salt stress: Importance of the arginine decarboxylase pathway in stress response. J. Exp. Bot. 2006, 57, 2589–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menéndez, A.B.; Calzadilla, P.I.; Sansberro, P.A.; Espasandin, F.D.; Gazquez, A.; Bordenave, C.D.; Maiale, S.J.; Rodríguez, A.A.; Maguire, V.G.; Campestre, M.P.; et al. Polyamines and Legumes: Joint Stories of Stress, Nitrogen Fixation and Environment. Front. Plant Sci. 2019, 10, 1415. [Google Scholar] [CrossRef] [PubMed]
- Poltronieri, P.; Bonsegna, S.; De Domenico, S.; Santino, A. Molecular mechanisms in plant abiotic stress response. Plant Abiotic Stress Response 2011, 48, 15–24. [Google Scholar] [CrossRef]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, 1–11. [Google Scholar] [CrossRef]
- Zhang, Z.; Mao, C.; Shi, Z.; Kou, X. The amino acid metabolic and carbohydrate metabolic pathway play important roles during salt-stress response in tomato. Front. Plant Sci. 2017, 8, 1231. [Google Scholar] [CrossRef] [Green Version]
- El-samad, H.A.; Barakat, N. The role of amino acids in improvement in salt tolerance of crop plants. J. Stress Physiol. Biochem. 2010, 6, 25–37. [Google Scholar]
- Conde, A.; Silva, P.; Agasse, A.; Conde, C.; Gerós, H. Mannitol transport and mannitol dehydrogenase activities are coordinated in olea Europaea under salt and osmotic stresses. Plant Cell Physiol. 2011, 52, 1766–1775. [Google Scholar] [CrossRef] [Green Version]
- Kaya, C.; Sonmez, O.; Aydemir, S.; Ashraf, M.; Dikilitas, M. Exogenous application of mannitol and thiourea regulates plant growth and oxidative stress responses in salt-stressed maize (Zea mays L.). J. Plant Interact. 2013, 8, 234–241. [Google Scholar] [CrossRef]
- Khalid, K.A.; Cai, W. The effects of mannitol and salinity stresses on growth and biochemical accumulations in lemon balm. Shengtai Xuebao/Acta Ecol. Sin. 2011, 31, 112–120. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, D.; Li, M.; Shi, L. Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS ONE 2016, 11, e159622. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Sun, T.; Li, S.; Xie, Y.; Song, X.; Wang, F.; Chen, L. Improved Salt Tolerance and Metabolomics Analysis of Synechococcus elongatus UTEX 2973 by Overexpressing Mrp Antiporters. Front. Bioeng. Biotechnol. 2020, 8, 500. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Chen, S.; Wang, Y. Advances in Understanding the Physiological and Molecular Responses of Sugar Beet to Salt Stress. Front. Plant Sci. 2019, 10, 1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemati, I.; Moradi, F.; Gholizadeh, S.; Esmaeili, M.A.; Bihamta, M.R. The effect of salinity stress on ions and soluble sugars distribution in leaves, leaf sheaths and roots of rice (Oryza sativa L.) seedlings. Plant, Soil Environ. 2011, 57, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Reza Amirjani, M. Effect of Salinity Stress on Growth, Sugar Content, Pigments and Enzyme Activity of Rice. Int. J. Bot. 2011, 7, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, A.; Sawada, H.; Shim, I.S.; Usui, K.; Fujihara, S. Effect of salt stress on physiological response and leaf polyamine content in NERICA rice seedlings. Plant Soil Environ. 2011, 57, 571–576. [Google Scholar] [CrossRef] [Green Version]
- Kiani, R.; Arzani, A.; Mirmohammady Maibody, S.A.M. Polyphenols, Flavonoids, and Antioxidant Activity Involved in Salt Tolerance in Wheat, Aegilops cylindrica and Their Amphidiploids. Front. Plant Sci. 2021, 12, 646221. [Google Scholar] [CrossRef] [PubMed]
- Sarker, U. Stress Hạn, Ảnh Hưởng Lên Sự Tạo Hợp Chất Thứ Cấp.Pdf. 2018. [Google Scholar]
- Linić, I.; Šamec, D.; Grúz, J.; Vujčić Bok, V.; Strnad, M.; Salopek-Sondi, B. Involvement of phenolic acids in short-term adaptation to salinity stress is species-specific among brassicaceae. Plants 2019, 8, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valifard, M.; Mohsenzadeh, S.; Kholdebarin, B.; Rowshan, V. Effects of salt stress on volatile compounds, total phenolic content and antioxidant activities of Salvia mirzayanii. S. Afr. J. Bot. 2014, 93, 92–97. [Google Scholar] [CrossRef] [Green Version]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Dias, M.C.; Freitas, H. Drought and Salinity Stress Responses and Microbe-Induced Tolerance in Plants. Front. Plant Sci. 2020, 11, 591911. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Hao, S.; Wang, Y.; Yan, Y.; Liu, Y.; Wang, J.; Chen, S. A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae 2021, 7, 132. [Google Scholar] [CrossRef]
- Khan, W.U.D.; Aziz, T.; Maqsood, M.A.; Farooq, M.; Abdullah, Y.; Ramzani, P.M.A.; Bilal, H.M. Silicon nutrition mitigates salinity stress in maize by modulating ion accumulation, photosynthesis, and antioxidants. Photosynthetica 2018, 56, 1047–1057. [Google Scholar] [CrossRef]
- Bekker, T.F.; Labuschagne, N.; Aveling, T.; Kaiser, C.; Regnier, T. Accumulation of total phenolics due to silicon application in roots of avocado trees infected with Phytophthora cinnamomi. S. Afr. Avocado Grow. Assoc. Yearb. 2007, 30, 57–64. [Google Scholar]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phoboo, S.; Sarkar, D.; Bhowmik, P.C.; Jha, P.K.; Shetty, K. Improving salinity resilience in Swertia chirayita clonal line with Lactobacillus plantarum. Can. J. Plant Sci. 2016, 96, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, B.; Liu, D.; Zou, C.; Wu, P.; Wang, Z.; Wang, Y.; Li, C. Transcriptomic and metabolomic analyses reveal mechanisms of adaptation to salinity in which carbon and nitrogen metabolism is altered in sugar beet roots. BMC Plant Biol. 2020, 20, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.S.; Persicke, M.; Elsayed, A.I.; Kalinowski, J.; Dietz, K. Metabolite profiling at the cellular and subcellular level reveals metabolites associated with salinity tolerance in sugar beet. J. Exp. Bot. 2017, 68, 5961–5976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, R. Salt-Tolerance Mechanisms in Plants; 2019; ISBN 9789353352134. [Google Scholar]
- Filippou, P.; Zarza, X.; Antoniou, C.; Obata, T.; Villarroel, C.A.; Ganopoulos, I.; Harokopos, V.; Gohari, G.; Aidinis, V.; Madesis, P.; et al. Systems biology reveals key tissue-specific metabolic and transcriptional signatures involved in the response of Medicago truncatula plant genotypes to salt stress. Comput. Struct. Biotechnol. J. 2021, 19, 2133–2147. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, D.; Liu, Q. Connections Between Amino Acid Metabolisms in Plants: Lysine as an Example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef] [PubMed]




| Pathways | Description | References |
|---|---|---|
| Glycolysis | Multistep reversible metabolic pathway for the production of pyruvate, a substrate for TCA cycle, from glucose | [124,125] |
| Pentose phosphate pathway | Branches from glucose-6-phosphate and ribose-5-phosphate, it provides substrates for oxidative defence | [126,127] |
| Tricarboxylic acid cycle | Central pathway for multiple enzyme-catalysed synthesis of organic acids from oxidation of pyruvate Provides energy to sustain anabolic and catabolic reactions involved in stress responses | [125,128] |
| Amino acid biosynthesis pathways | Several branched pathways in which amino acids are synthesized from their precursors, | [129,130] |
| Linoleic and other fatty acids pathways | Synthesis of lipids with roles in stress signalling cascades and precursors for phytohormones synthesis | [131,132] |
| Shikimic pathway | Synthesis of shikimic acid (precursor for phenylalanine synthesis) from the downstream products of the pentose phosphate pathway, erythrose-4-phosphate and phosphoenol | [98,133] |
| Phenylpropanoid pathway | The primary pathway responsible for production of all phenolics from phenylalanine | [69,134] |
| Flavonoids biosynthetic pathway | An offramp from the phenylpropanoid pathway, produces a range of flavonoids from 4-coumaroyl-CoA | [135,136] |
| Polyamine biosynthesis pathways | Synthesis of polyamines (spermine, spermidine, and putrescene) from ornithine, methionine, and arginine | [137,138] |
| Metabolite Group | Stress-Responsive Roles | Plant Species | References |
|---|---|---|---|
| PRIMARY METABOLITES | |||
| Amino acids | ROS scavenging (proline), protein stabilisation and synthesis, redox control | Tomato, Maize and beans | [141,142] |
| Polyols | Protection of photosynthesis systems, ROS scavenging, protein stabilisation | Olea europaea, maize, Melissa officinalis | [143,144,145] |
| Organic acids | Energy production, signalling molecules, antioxidant activities | Rice, soybean | [146,147] |
| Sugars | Signalling molecules, carbon energy reserve, maintenance of redox homeostasis, osmoprotectants | Beta vulgaris, Oryza sativa | [148,149,150] |
| SECONDARY METABOLITES | |||
| Polyamines | Activation of antioxidant enzymes, regulation of ion channels activity, protein and membrane stabilisation | Legumes, rice | [138,151] |
| Flavonoids | Radical scavenging, inhibition of pro-oxidant enzymes, stimulate ROS response genes | Aegilops cylindrica, Amaranthus tricolor | [152,153] |
| Phenolic acids | Hormonal regulation, antioxidant activity, photosynthetic activity, improve nutrient uptake | Brassica crops, Salvia mirzayanii, Thymus species | [11,136,154] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chele, K.H.; Tinte, M.M.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective. Metabolites 2021, 11, 724. https://doi.org/10.3390/metabo11110724
Chele KH, Tinte MM, Piater LA, Dubery IA, Tugizimana F. Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective. Metabolites. 2021; 11(11):724. https://doi.org/10.3390/metabo11110724
Chicago/Turabian StyleChele, Kekeletso H., Morena M. Tinte, Lizelle A. Piater, Ian A. Dubery, and Fidele Tugizimana. 2021. "Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective" Metabolites 11, no. 11: 724. https://doi.org/10.3390/metabo11110724