Involvement of Dmp1 in the Precise Regulation of Hair Bundle Formation in the Developing Cochlea
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Immunohistochemistry
2.3. Electron Microscopy
2.4. Quantitative qPCR
2.5. Western Blotting
2.6. RNA Sequencing and Bioinformatic Analysis
2.7. Phenotypic and Statistical Analysis
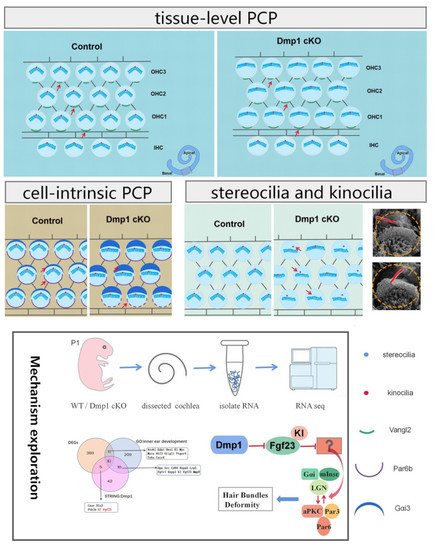
3. Results
3.1. Dmp1 Expression in Developing HCs and Conditional Inactivation of Dmp1 in the Cochlea
3.2. Dmp1 Deficiency Leads to Stereociliary Bundle Deformity but Not the Loss of HCs
3.3. Misplaced or Occasionally Absent Kinocilia in Dmp1-Deficient Cochlea
3.4. Dmp1 Deficiency Affects Intrinsic Cell Polarity Rather than Planar Cell Polarity
3.5. Transcriptomic Changes in Dmp1-Deficient Cochlea
4. Discussion
4.1. Roles and Studies of Dmp1 in the Mouse Ear
4.2. Abnormalities in Stereociliary Morphology and a Misorientation of Kinocilia in Dmp1 cKD Mice
4.3. Dmp1 Deficiency Affects the Cell-Intrinsic Polarity of HCs
4.4. Transcriptomic Changes in the Cochlea of Dmp1 cKD Mice
4.5. The Normal Hearing Function in Adult Dmp1 cKD Mice
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morton, C.C.; Nance, W.E. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006, 354, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- Jackler, R.K.; Luxford, W.M.; House, W.F. Congenital malformations of the inner ear: A classification based on embryogenesis. Laryngoscope 1987, 97 (Suppl. S40), 2–14. [Google Scholar] [CrossRef]
- Marazita, M.L.; Ploughman, L.M.; Rawlings, B.; Remington, E.; Arnos, K.S.; Nance, W.E. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am. J. Med. Genet. 1993, 46, 486–491. [Google Scholar] [CrossRef]
- Lenz, D.R.; Avraham, K.B. Hereditary hearing loss: From human mutation to mechanism. Hear. Res. 2011, 281, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Raphael, Y.; Altschuler, R.A. Structure and innervation of the cochlea. Brain Res. Bull. 2003, 60, 397–422. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, E.; Krupin, A.; Kelley, M.W. Cellular growth and rearrangement during the development of the mammalian organ of Corti. Dev. Dyn. 2004, 229, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, P.; Müller, U. Sensing sound: Molecules that orchestrate mechanotransduction by hair cells. Trends Neurosci. 2012, 35, 220–229. [Google Scholar] [CrossRef]
- Tarchini, B.; Lu, X. New insights into regulation and function of planar polarity in the inner ear. Neurosci. Lett. 2019, 709, 134373. [Google Scholar] [CrossRef]
- Ogbureke, K.U.; Fisher, L.W. Renal expression of SIBLING proteins and their partner matrix metalloproteinases (MMPs). Kidney Int. 2005, 68, 155–166. [Google Scholar] [CrossRef]
- George, A.; Silberstein, R.; Veis, A. In situ hybridization shows Dmp1 (AG1) to be a developmentally regulated dentin-specific protein produced by mature odontoblasts. Connect. Tissue Res. 1995, 33, 67–72. [Google Scholar] [CrossRef]
- Toyosawa, S.; Shintani, S.; Fujiwara, T.; Ooshima, T.; Sato, A.; Ijuhin, N.; Komori, T. Dentin matrix protein 1 is predominantly expressed in chicken and rat osteocytes but not in osteoblasts. J. Bone Miner. Res. 2001, 16, 2017–2026. [Google Scholar] [CrossRef]
- Qin, C.; D’Souza, R.; Feng, J.Q. Dentin matrix protein 1 (DMP1): New and important roles for biomineralization and phosphate homeostasis. J. Dent. Res. 2007, 86, 1134–1141. [Google Scholar] [CrossRef]
- Terasawa, M.; Shimokawa, R.; Terashima, T.; Ohya, K.; Takagi, Y.; Shimokawa, H. Expression of dentin matrix protein 1 (DMP1) in nonmineralized tissues. J. Bone Miner. Metab. 2004, 22, 430–438. [Google Scholar] [CrossRef]
- Jing, B.; Zhang, C.; Liu, X.; Zhou, L.; Liu, J.; Yao, Y.; Yu, J.; Weng, Y.; Pan, M.; Liu, J.; et al. Glycosylation of dentin matrix protein 1 is a novel key element for astrocyte maturation and BBB integrity. Protein Cell 2018, 9, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Anunobi, C.C.; Koli, K.; Saxena, G.; Banjo, A.A.; Ogbureke, K.U. Expression of the SIBLINGs and their MMP partners in human benign and malignant prostate neoplasms. Oncotarget 2016, 7, 48038–48049. [Google Scholar] [CrossRef] [PubMed]
- Mäkitie, O.; Pereira, R.C.; Kaitila, I.; Turan, S.; Bastepe, M.; Laine, T.; Kröger, H.; Cole, W.G.; Jüppner, H. Long-term clinical outcome and carrier phenotype in autosomal recessive hypophosphatemia caused by a novel DMP1 mutation. J. Bone Miner. Res. 2010, 25, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.; Kane, R.; Valentine, J. Impaired hearing in X-linked hypophosphataemic (vitamin-D-resistant) osteomalacia. Ann. Intern. Med. 1984, 100, 230–232. [Google Scholar] [CrossRef]
- Meister, M.; Johnson, A.; Popelka, G.R.; Kim, G.S.; Whyte, M.P. Audiologic findings in young patients with hypophosphatemic bone disease. Ann. Otol. Rhinol. Laryngol. 1986, 95 Pt 1, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Weir, N. Sensorineural deafness associated with recessive hypophosphataemic rickets. J. Laryngol. Otol. 1977, 91, 717–722. [Google Scholar] [CrossRef]
- O’Malley, S.; Ramsden, R.T.; Latif, A.; Kane, R.; Davies, M. Electrocochleographic changes in the hearing loss associated with X-linked hypophosphataemic osteomalacia. Acta Otolaryngol. 1985, 100, 13–18. [Google Scholar] [CrossRef]
- Li, W.X.; Peng, H.; Yang, L.; Hao, Q.Q.; Sun, W.; Ji, F.; Guo, W.W.; Yang, S.M. Familial nonsyndromic hearing loss with incomplete partition type II caused by novel DSPP gene mutations. Acta Otolaryngol. 2018, 138, 685–690. [Google Scholar] [CrossRef]
- Xiao, S.; Yu, C.; Chou, X.; Yuan, W.; Wang, Y.; Bu, L.; Fu, G.; Qian, M.; Yang, J.; Shi, Y.; et al. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet. 2001, 27, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Nam, S.H.; Jang, K.T.; Lee, S.H.; Kim, C.C.; Hahn, S.H.; Hu, J.C.; Simmer, J.P. A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum. Genet. 2004, 115, 248–254. [Google Scholar] [CrossRef]
- Zhang, S.; Wan, H.; Wang, P.; Liu, M.; Li, G.; Zhang, C.; Sun, Y. Extracellular matrix protein DMP1 suppresses osteogenic differentiation of Mesenchymal Stem Cells. Biochem. Biophys. Res. Commun. 2018, 501, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Hébert, J.M.; McConnell, S.K. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 2000, 222, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Qian, X.Q.; Yang, X.Y.; Jiang, T.; Wang, Y.M.; Lyu, J.H.; Chi, F.L.; Chen, P.; Ren, D.D. Rab11a Regulates the Development of Cilia and Establishment of Planar Cell Polarity in Mammalian Vestibular Hair Cells. Front. Mol. Neurosci. 2021, 14, 762916. [Google Scholar]
- Lv, K.; Huang, H.; Yi, X.; Chertoff, M.E.; Li, C.; Yuan, B.; Hinton, R.J.; Feng, J.Q. A novel auditory ossicles membrane and the development of conductive hearing loss in Dmp1-null mice. Bone 2017, 103, 39–46. [Google Scholar] [CrossRef]
- Lv, K.; Huang, H.; Lu, Y.; Qin, C.; Li, Z.; Feng, J.Q. Circling behavior developed in Dmp1 null mice is due to bone defects in the vestibular apparatus. Int. J. Biol. Sci. 2010, 6, 537–545. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Y.; Lundberg, Y.W. Spatiotemporal differences in otoconial gene expression. Genesis 2016, 54, 613–625. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, H.; Yang, H.; Zhao, X.; Lovas, S.; Lundberg, Y.W. Expression, functional, and structural analysis of proteins critical for otoconia development. Dev. Dyn. 2010, 239, 2659–2673. [Google Scholar] [CrossRef]
- Maoiléidigh, D.Ó.; Ricci, A.J. A Bundle of Mechanisms: Inner-Ear Hair-Cell Mechanotransduction. Trends Neurosci. 2019, 42, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Schwander, M.; Kachar, B.; Muller, U. Review series: The cell biology of hearing. J. Cell Biol. 2010, 190, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.J.; May-Simera, H.; Eichers, E.R.; Kai, M.; Hill, J.; Jagger, D.J.; Leitch, C.C.; Chapple, J.P.; Munro, P.M.; Fisher, S.; et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat. Genet. 2005, 37, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Chatterjee, B.; Francis, D.; Yu, Q.; SanAgustin, J.T.; Francis, R.; Tansey, T.; Henry, C.; Wang, B.; Lemley, B.; et al. Disruption of Mks1 localization to the mother centriole causes cilia defects and developmental malformations in Meckel-Gruber syndrome. Dis. Model. Mech. 2011, 4, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Sipe, C.W.; Lu, X. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development 2011, 138, 3441–3449. [Google Scholar] [CrossRef]
- Jones, C.; Roper, V.C.; Foucher, I.; Qian, D.; Banizs, B.; Petit, C.; Yoder, B.K.; Chen, P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat. Genet. 2008, 40, 69–77. [Google Scholar] [CrossRef]
- Webb, S.W.; Grillet, N.; Andrade, L.R.; Xiong, W.; Swarthout, L.; Della Santina, C.C.; Kachar, B.; Muller, U. Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 2011, 138, 1607–1617. [Google Scholar] [CrossRef]
- Li, R.; Gundersen, G.G. Beyond polymer polarity: How the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 2008, 9, 860–873. [Google Scholar] [CrossRef]
- Sipe, C.W.; Liu, L.; Lee, J.; Grimsley-Myers, C.; Lu, X. Lis1 mediates planar polarity of auditory hair cells through regulation of microtubule organization. Development 2013, 140, 1785–1795. [Google Scholar] [CrossRef]
- Ezan, J.; Lasvaux, L.; Gezer, A.; Novakovic, A.; May-Simera, H.; Belotti, E.; Lhoumeau, A.C.; Birnbaumer, L.; Beer-Hammer, S.; Borg, J.P.; et al. Primary cilium migration depends on G-protein signalling control of subapical cytoskeleton. Nat. Cell Biol. 2013, 15, 1107–1115. [Google Scholar] [CrossRef]
- Montcouquiol, M.; Rachel, R.A.; Lanford, P.J.; Copeland, N.G.; Jenkins, N.A.; Kelley, M.W. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 2003, 423, 173–177. [Google Scholar] [CrossRef]
- Song, H.; Hu, J.; Chen, W.; Elliott, G.; Andre, P.; Gao, B.; Yang, Y. Planar cell polarity breaks bilateral symmetry by controlling ciliary positioning. Nature 2010, 466, 378–382. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, N.; Nathans, J. The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J. Neurosci. 2006, 26, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
- Grimsley-Myers, C.M.; Sipe, C.W.; Geleoc, G.S.; Lu, X. The small GTPase Rac1 regulates auditory hair cell morphogenesis. J. Neurosci. 2009, 29, 15859–15869. [Google Scholar] [CrossRef] [PubMed]
- Landin Malt, A.; Dailey, Z.; Holbrook-Rasmussen, J.; Zheng, Y.; Hogan, A.; Du, Q.; Lu, X. Par3 is essential for the establishment of planar cell polarity of inner ear hair cells. Proc. Natl. Acad. Sci. USA 2019, 116, 4999–5008. [Google Scholar] [CrossRef] [PubMed]
- Tarchini, B.; Jolicoeur, C.; Cayouette, M. A molecular blueprint at the apical surface establishes planar asymmetry in cochlear hair cells. Dev. Cell 2013, 27, 88–102. [Google Scholar] [CrossRef]
- Nguyen-Ngoc, T.; Afshar, K.; Gönczy, P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 2007, 9, 1294–1302. [Google Scholar] [CrossRef]
- Beer-Hammer, S.; Lee, S.C.; Mauriac, S.A.; Leiss, V.; Groh, I.A.M.; Novakovic, A.; Piekorz, R.P.; Bucher, K.; Chen, C.; Ni, K.; et al. Gαi Proteins are Indispensable for Hearing. Cell Physiol. Biochem. 2018, 47, 1509–1532. [Google Scholar] [CrossRef]
- Durgan, J.; Kaji, N.; Jin, D.; Hall, A. Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J. Biol. Chem. 2011, 286, 12461–12474. [Google Scholar] [CrossRef]
- Joberty, G.; Petersen, C.; Gao, L.; Macara, I.G. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2000, 2, 531–539. [Google Scholar] [CrossRef]
- Petronczki, M.; Knoblich, J.A. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 2001, 3, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.Y.; Soares, V.Y.; Kristiansen, A.G.; Stankovic, K.M. Activation of TRAIL-DR5 pathway promotes sensorineural degeneration in the inner ear. Aging Cell 2016, 15, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Lee, M.H.; Kang, S.U.; Hwang, H.S.; Park, K.; Choung, Y.H.; Kim, C.H. Nitric oxide mediates TNF-α-induced apoptosis in the auditory cell line. Laryngoscope 2012, 122, 2256–2264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zheng, H.; Pyykko, I.; Zou, J. The TLR-4/NF-κB signaling pathway activation in cochlear inflammation of rats with noise-induced hearing loss. Hear. Res. 2019, 379, 59–68. [Google Scholar] [CrossRef]
- Li, X. The FGF metabolic axis. Front. Med. 2019, 13, 511–530. [Google Scholar] [CrossRef]
- Rodríguez, M. FGF23: Is It Another Biomarker for Phosphate-Calcium Metabolism? Adv. Ther. 2020, 37 (Suppl. 2), 73–79. [Google Scholar] [CrossRef]
- Urakawa, Y.; Yamazaki, T.; Shimada, K.; Iijima, H.; Hasegawa, K.; Okawa, K.; Fujita, T.; Fukumoto, S.; Yamashita, T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 2006, 444, 770–774. [Google Scholar] [CrossRef]
- Kurosu, H.; Ogawa, Y.; Miyoshi, M.; Yamamoto, M.; Nandi, A.; Rosenblatt, K.P.; Schiavi, S.; Hu, M.C.; Moe, O.W.; Kuro-o, M. Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 2006, 281, 6120–6123. [Google Scholar] [CrossRef]
- Quarles, L.D. Endocrine functions of bone in mineral metabolism regulation. J. Clin. Investig. 2008, 118, 3820–3828. [Google Scholar] [CrossRef]
- Kamemori, M.; Ohyama, Y.; Kurabayashi, M.; Takahashi, K.; Nagai, R.; Furuya, N. Expression of klotho protein in the inner ear. Hear. Res. 2002, 171, 103–110. [Google Scholar] [CrossRef]
- Lysaght, A.C.; Yuan, Q.; Fan, Y.; Kalwani, N.; Caruso, P.; Cunnane, M.; Lanske, B.; Stanković, K.M. FGF23 deficiency leads to mixed hearing loss and middle ear malformation in mice. PLoS ONE 2014, 9, e107681. [Google Scholar] [CrossRef] [PubMed]
- Copley, C.O.; Duncan, J.S.; Liu, C.; Cheng, H.; Deans, M.R. Postnatal refinement of auditory hair cell planar polarity deficits occurs in the absence of Vangl2. J. Neurosci. 2013, 33, 14001–14016. [Google Scholar] [CrossRef] [PubMed]








| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Cxcr4 | CCATGGAACCGATCAGTGTGAGTA | TTGTCCGTCATGCTCCTTAGCTTC |
| Wnt3 | CACAACACGAGGACGGAGA | AATCTACCCCTTCCCAGTGC |
| Atoh1 | GTA AGG AGA AGC GGC TGT | AGC CAA GCT CGT CCA CTA |
| Hes1 | CCAGCCAGTGTCAACACGA | AATGCCGGGAGCTATCTTTCT |
| Ptger4 | ACCATTCCTAGATCGAACCGT | CACCACCCCGAAGATGAACAT |
| Gapdh | GCAAGGACACTGAGCAAGA | GGATGGAAATTGTGAGGGAG |
| Gene Symbol | log2FoldChange | p Value | FDR | Up or Down |
|---|---|---|---|---|
| Gpha2 | 3.731 | 0.00002 | 0.009 | Down |
| Actc1 | 3.702 | 0.00002 | 0.010 | Down |
| Olig2 | 3.612 | 0.00002 | 0.010 | Up |
| Gm3558 | 3.561 | 0.00004 | 0.016 | Down |
| Lgals1-ps2 | 3.465 | 0.00016 | 0.035 | Down |
| Gdf1 | 2.874 | 0.00003 | 0.011 | Up |
| Il11ra2 | 2.433 | 0.00000 | 0.000 | Up |
| Atoh1 | 2.342 | 0.00003 | 0.013 | Down |
| Oasl1 | 2.266 | 0.00019 | 0.039 | Down |
| Eid3 | 2.054 | 0.00001 | 0.006 | Up |
| Bdkrb2 | 2.011 | 0.00000 | 0.003 | Down |
| Cxcr4 | 2.010 | 0.00000 | 0.000 | Down |
| Sox18 | 2.000 | 0.00001 | 0.005 | Down |
| Slc16a3 | 1.782 | 0.00007 | 0.020 | Down |
| Ptger4 | 1.776 | 0.00000 | 0.003 | Down |
| Ifit3 | 1.725 | 0.00016 | 0.035 | Down |
| Zfp599 | 1.685 | 0.00005 | 0.016 | Down |
| Clec2d | 1.640 | 0.00000 | 0.002 | Down |
| Hes1 | 1.549 | 0.00010 | 0.025 | Down |
| Zbtb4 | 1.537 | 0.00000 | 0.000 | Up |
| Cnih3 | 1.529 | 0.00025 | 0.047 | Up |
| Cited2 | 1.527 | 0.00000 | 0.000 | Down |
| Elf3 | 1.430 | 0.00022 | 0.042 | Down |
| Ntf3 | 1.371 | 0.00001 | 0.005 | Down |
| Fzd8 | 1.342 | 0.00004 | 0.016 | Down |
| Myc | 1.293 | 0.00000 | 0.000 | Down |
| Ccnd3 | 1.151 | 0.00001 | 0.006 | Down |
| Rtp4 | 1.098 | 0.00016 | 0.035 | Down |
| Zfp84 | 1.097 | 0.00003 | 0.014 | Down |
| Wnt3 | 1.096 | 0.00007 | 0.020 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Lyu, J.; Qian, X.; Chen, B.; Sun, H.; Luo, W.; Chi, F.; Li, H.; Ren, D. Involvement of Dmp1 in the Precise Regulation of Hair Bundle Formation in the Developing Cochlea. Biology 2023, 12, 625. https://doi.org/10.3390/biology12040625
Wang Y, Lyu J, Qian X, Chen B, Sun H, Luo W, Chi F, Li H, Ren D. Involvement of Dmp1 in the Precise Regulation of Hair Bundle Formation in the Developing Cochlea. Biology. 2023; 12(4):625. https://doi.org/10.3390/biology12040625
Chicago/Turabian StyleWang, Yanmei, Jihan Lyu, Xiaoqing Qian, Binjun Chen, Haojie Sun, Wenwei Luo, Fanglu Chi, Hongzhe Li, and Dongdong Ren. 2023. "Involvement of Dmp1 in the Precise Regulation of Hair Bundle Formation in the Developing Cochlea" Biology 12, no. 4: 625. https://doi.org/10.3390/biology12040625