Replication of Integrative Data Analysis for Adipose Tissue Dysfunction, Low-Grade Inflammation, Postprandial Responses and OMICs Signatures in Symptom-Free Adults
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
3. Results
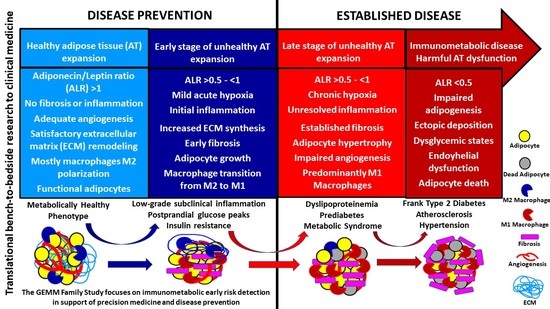
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stefan, N.; Haring, H.U.; Hu, F.B.; Schulze, M.B. Metabolically healthy obesity: Epidemiology, mechanisms, and clinical implications. Lancet Diabetes Endocrinol. 2013, 1, 152–162. [Google Scholar] [CrossRef]
- Virtue, S.; Vidal-Puig, A. It’s not how fat you are, it’s what you do with it that counts. PLoS Biol. 2008, 6, e237. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, L.; Mollica, M.P.; Lombardi, A.; Cavaliere, G.; Gifuni, G.; Barletta, A. From chronic overnutrition to insulin resistance: The role of fat-storing capacity and inflammation. Nutr. Metab. Cardiovasc. Dis. 2009, 19, 146–152. [Google Scholar] [CrossRef]
- Stefan, N.; Schick, F.; Haring, H.U. Causes, Characteristics, and Consequences of Metabolically Unhealthy Normal Weight in Humans. Cell Metab. 2017, 26, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, C.; Cataldo, L.R.; Galgani, J.; Parada, J.; Santos, J.L. Leptin/Adiponectin Ratios Using Either Total Or High-Molecular-Weight Adiponectin as Biomarkers of Systemic Insulin Sensitivity in Normoglycemic Women. J. Diabetes Res. 2017, 2017, 9031079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruhbeck, G.; Catalan, V.; Rodriguez, A.; Ramirez, B.; Becerril, S.; Salvador, J.; Colina, I.; Gomez-Ambrosi, J. Adiponectin-Leptin Ratio Is a Functional Biomarker of Adipose Tissue Inflammation. Nutrients 2019, 11, 454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruhbeck, G.; Catalan, V.; Rodriguez, A.; Ramirez, B.; Becerril, S.; Salvador, J.; Portincasa, P.; Colina, I.; Gomez-Ambrosi, J. Involvement of the leptin-adiponectin axis in inflammation and oxidative stress in the metabolic syndrome. Sci. Rep. 2017, 7, 6619. [Google Scholar] [CrossRef] [PubMed]
- Fruhbeck, G.; Catalan, V.; Rodriguez, A.; Gomez-Ambrosi, J. Adiponectin-leptin ratio: A promising index to estimate adipose tissue dysfunction. Relation with obesity-associated cardiometabolic risk. Adipocyte 2018, 7, 57–62. [Google Scholar] [CrossRef]
- Aguilar-Salinas, C.A.; Velazquez Monroy, O.; Gomez-Perez, F.J.; Gonzalez Chavez, A.; Esqueda, A.L.; Molina Cuevas, V.; Rull-Rodrigo, J.A.; Tapia Conyer, R. Characteristics of patients with type 2 diabetes in Mexico: Results from a large population-based nationwide survey. Diabetes Care 2003, 26, 2021–2026. [Google Scholar] [CrossRef] [Green Version]
- Harris, M.I.; Flegal, K.M.; Cowie, C.C.; Eberhardt, M.S.; Goldstein, D.E.; Little, R.R.; Wiedmeyer, H.M.; Byrd-Holt, D.D. Prevalence of diabetes, impaired fasting glucose, and impaired glucose tolerance in U.S. adults. The Third National Health and Nutrition Examination Survey, 1988–1994. Diabetes Care 1998, 21, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Morales, L.S.; Flores, Y.N.; Leng, M.; Sportiche, N.; Gallegos-Carrillo, K.; Salmeron, J. Risk factors for cardiovascular disease among Mexican-American adults in the United States and Mexico: A comparative study. Salud Publica Mex. 2014, 56, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Barquera, S.; Campos, I.; Rivera, J.A. Mexico attempts to tackle obesity: The process, results, push backs and future challenges. Obes. Rev. 2013, 14, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Bastarrachea, R.A.; Laviada-Molina, H.A.; Nava-Gonzalez, E.J.; Leal-Berumen, I.; Escudero-Lourdes, C.; Escalante-Araiza, F.; Peschard, V.G.; Veloz-Garza, R.A.; Haack, K.; Martinez-Hernandez, A.; et al. Deep Multi-OMICs and Multi-Tissue Characterization in a Pre- and Postprandial State in Human Volunteers: The GEMM Family Study Research Design. Genes 2018, 9, 532. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Ayala, E.; Gallegos-Cabrales, E.C.; Gonzalez-Lopez, L.; Laviada-Molina, H.A.; Salinas-Osornio, R.A.; Nava-Gonzalez, E.J.; Leal-Berumen, I.; Escudero-Lourdes, C.; Escalante-Araiza, F.; Buenfil-Rello, F.A.; et al. Towards precision medicine: Defining and characterizing adipose tissue dysfunction to identify early immunometabolic risk in symptom-free adults from the GEMM family study. Adipocyte 2020, 9, 153–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, B.A.; Caskey, C.T.; Brar, P.; Dec, E.; Karow, D.S.; Kahn, A.M.; Hou, Y.C.; Shah, N.; Boeldt, D.; Coughlin, E.; et al. Precision medicine screening using whole-genome sequencing and advanced imaging to identify disease risk in adults. Proc. Natl. Acad. Sci. USA 2018, 115, 3686–3691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San-Cristobal, R.; Navas-Carretero, S.; Martínez-González, M.; Ordovas, J.M.; Martínez, J.A. Contribution of macronutrients to obesity: Implications for precision nutrition. Nat. Rev. Endocrinol. 2020, 16, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Olendzki, B.C.; Pagoto, S.L.; Hurley, T.G.; Magner, R.P.; Ockene, I.S.; Schneider, K.L.; Merriam, P.A.; Hébert, J.R. Number of 24-hour diet recalls needed to estimate energy intake. Ann. Epidemiol. 2009, 19, 553–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, S.; Keitel-Korndorfer, A.; Herfurth-Majstorovic, K.; Wendt, V.; Klein, A.M.; von Klitzing, K.; Grube, M. Recruitment strategies in a prospective longitudinal family study on parents with obesity and their toddlers. BMC Public Health 2017, 17, 145. [Google Scholar] [CrossRef] [Green Version]
- Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Frost, P.A.; Chen, S.; Rodriguez-Ayala, E.; Laviada-Molina, H.A.; Vaquera, Z.; Gaytan-Saucedo, J.F.; Li, W.-H.; Haack, K.; Grayburn, P.A.; Sayers, K.; et al. Research methodology for in vivo measurements of resting energy expenditure, daily body temperature, metabolic heat and non-viral tissue-specific gene therapy in baboons. Res. Vet. Sci. 2020, 133, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Wang, M.; Wang, C.; Han, R.H.; Han, X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016, 61, 83–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Lei, X.; Kujawa, S.A.; Bolu, N.; Zhao, H.; Wang, C. Identification of core gene in obese type 2 diabetes patients using bioinformatics analysis. Adipocyte 2021, 10, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Du, T.; Zhang, J.; Yuan, G.; Zhang, M.; Zhou, X.; Liu, Z.; Sun, X.; Yu, X. Nontraditional risk factors for cardiovascular disease and visceral adiposity index among different body size phenotypes. Nutr. Metab. Cardiovasc. Dis. 2015, 25, 100–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahalle, N.P.; Garg, M.K.; Kulkarni, M.V.; Naik, S.S. Differences in traditional and non-traditional risk factors with special reference to nutritional factors in patients with coronary artery disease with or without diabetes mellitus. Indian J. Endocrinol. Metab. 2013, 17, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Furugen, M.; Saitoh, S.; Ohnishi, H.; Akasaka, H.; Mitsumata, K.; Chiba, M.; Furukawa, T.; Miyazaki, Y.; Shimamoto, K.; Miura, T. Matsuda-DeFronzo insulin sensitivity index is a better predictor than HOMA-IR of hypertension in Japanese: The Tanno-Sobetsu study. J. Hum. Hypertens. 2012, 26, 325–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietzner, M.; Kaul, A.; Henning, A.K.; Kastenmuller, G.; Artati, A.; Lerch, M.M.; Adamski, J.; Nauck, M.; Friedrich, N. Comprehensive metabolic profiling of chronic low-grade inflammation among generally healthy individuals. BMC Med. 2017, 15, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacobini, C.; Pugliese, G.; Blasetti Fantauzzi, C.; Federici, M.; Menini, S. Metabolically healthy versus metabolically unhealthy obesity. Metabolism 2019, 92, 51–60. [Google Scholar] [CrossRef]
- Smith, G.I.; Mittendorfer, B.; Klein, S. Metabolically healthy obesity: Facts and fantasies. J. Clin. Investig. 2019, 129, 3978–3989. [Google Scholar] [CrossRef] [Green Version]
- Mechanick, J.I.; Garber, A.J.; Grunberger, G.; Handelsman, Y.; Garvey, W.T. Dysglycemia-based chronic disease: An american association of clinical endocrinologists position statement. Endocr. Pract. 2018, 24, 995–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, M.L. Prediabetes: Beyond the Borderline. Nurs. Clin. N. Am. 2017, 52, 665–677. [Google Scholar] [CrossRef]
- Misra, A.; Wasir, J.S.; Vikram, N.K. Waist circumference criteria for the diagnosis of abdominal obesity are not applicable uniformly to all populations and ethnic groups. Nutrition 2005, 21, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.Q.; Li, Q.; Rentfro, A.R.; Fisher-Hoch, S.P.; McCormick, J.B. The definition of insulin resistance using HOMA-IR for Americans of Mexican descent using machine learning. PLoS ONE 2011, 6, e21041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Yang, K.; Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012, 31, 134–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagheri, M.; Farzadfar, F.; Qi, L.; Yekaninejad, M.S.; Chamari, M.; Zeleznik, O.A.; Kalantar, Z.; Ebrahimi, Z.; Sheidaie, A.; Koletzko, B.; et al. Obesity-Related Metabolomic Profiles and Discrimination of Metabolically Unhealthy Obesity. J. Proteome Res. 2018, 17, 1452–1462. [Google Scholar] [CrossRef]
- Caleyachetty, R.; Thomas, G.N.; Toulis, K.A.; Mohammed, N.; Gokhale, K.M.; Balachandran, K.; Nirantharakumar, K. Metabolically Healthy Obese and Incident Cardiovascular Disease Events among 3.5 Million Men and Women. J. Am. Coll. Cardiol. 2017, 70, 1429–1437. [Google Scholar] [CrossRef]
- Mathew, H.; Farr, O.M.; Mantzoros, C.S. Metabolic health and weight: Understanding metabolically unhealthy normal weight or metabolically healthy obese patients. Metabolism 2016, 65, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruderman, N.; Chisholm, D.; Pi-Sunyer, X.; Schneider, S. The metabolically obese, normal-weight individual revisited. Diabetes 1998, 47, 699–713. [Google Scholar] [CrossRef]
- Shea, J.L.; King, M.T.; Yi, Y.; Gulliver, W.; Sun, G. Body fat percentage is associated with cardiometabolic dysregulation in BMI-defined normal weight subjects. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Roberson, L.L.; Aneni, E.C.; Maziak, W.; Agatston, A.; Feldman, T.; Rouseff, M.; Tran, T.; Blaha, M.J.; Santos, R.D.; Sposito, A.; et al. Beyond BMI: The “Metabolically healthy obese” phenotype & its association with clinical/subclinical cardiovascular disease and all-cause mortality—A systematic review. BMC Public Health 2014, 14, 14. [Google Scholar]
- Franzosi, M.G. Should we continue to use BMI as a cardiovascular risk factor? Lancet 2006, 368, 624–625. [Google Scholar] [CrossRef]
- Crewe, C.; An, Y.A.; Scherer, P.E. The ominous triad of adipose tissue dysfunction: Inflammation, fibrosis, and impaired angiogenesis. J. Clin. Investig. 2017, 127, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef] [PubMed]
- Despres, J.P. Body fat distribution and risk of cardiovascular disease: An update. Circulation 2012, 126, 1301–1313. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.S.; Evans, C.V.; Johnson, E.; Redmond, N.; Coppola, E.L.; Smith, N. Nontraditional Risk Factors in Cardiovascular Disease Risk Assessment: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2018, 320, 281–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balagopal, P.B.; de Ferranti, S.D.; Cook, S.; Daniels, S.R.; Gidding, S.S.; Hayman, L.L.; McCrindle, B.W.; Mietus-Snyder, M.L.; Steinberger, J. Nontraditional risk factors and biomarkers for cardiovascular disease: Mechanistic, research, and clinical considerations for youth: A scientific statement from the American Heart Association. Circulation 2011, 123, 2749–2769. [Google Scholar] [CrossRef] [Green Version]
- Frühbeck, G.; Gómez-Ambrosi, J. Modulation of the leptin-induced white adipose tissue lipolysis by nitric oxide. Cell Signal 2001, 13, 827–833. [Google Scholar] [CrossRef]
- Asano, T.; Fujishiro, M.; Kushiyama, A.; Nakatsu, Y.; Yoneda, M.; Kamata, H.; Sakoda, H. Role of phosphatidylinositol 3-kinase activation on insulin action and its alteration in diabetic conditions. Biol. Pharm. Bull. 2007, 30, 1610–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, E.; Makrecka-Kuka, M.; Vilks, K.; Volska, K.; Sevostjanovs, E.; Grinberga, S.; Zarkova-Malkova, O.; Dambrova, M.; Liepinsh, E. Decreases in Circulating Concentrations of Long-Chain Acylcarnitines and Free Fatty Acids During the Glucose Tolerance Test Represent Tissue-Specific Insulin Sensitivity. Front. Endocrinol. 2019, 10, 870. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.O.; Koutsari, C.; Blachnio-Zabielska, A.U.; Hames, K.C.; Jensen, M.D. Effects of meal ingestion on intramyocellular ceramide concentrations and fractional de novo synthesis in humans. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E105–E114. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.E.; Parks, E.J. Postprandial metabolism of meal triglyceride in humans. Biochim. Biophys. Acta 2012, 1821, 721–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, U.A.; Fitch, M.D.; Beyl, R.A.; Hellerstein, M.K.; Ravussin, E. Differences in In Vivo Cellular Kinetics in Abdominal and Femoral Subcutaneous Adipose Tissue in Women. Diabetes 2016, 65, 1642–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahmad, H.F.; Daouk, R.; Azar, J.; Sapudom, J.; Teo, J.C.M.; Abou-Kheir, W.; Al-Sayegh, M. Modeling Adipogenesis: Current and Future Perspective. Cells 2020, 9, 2326. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; Krishnaswami, S.; Harris, T.B.; Katsiaras, A.; Kritchevsky, S.B.; Simonsick, E.M.; Nevitt, M.; Holvoet, P.; Newman, A.B. Obesity, regional body fat distribution, and the metabolic syndrome in older men and women. Arch. Intern. Med. 2005, 165, 777–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Gerhold, K.; Mayers, J.R.; Wiest, M.M.; Watkins, S.M.; Hotamisligil, G.S. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 2008, 134, 933–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefan, N.; Kantartzis, K.; Celebi, N.; Staiger, H.; Machann, J.; Schick, F.; Cegan, A.; Elcnerova, M.; Schleicher, E.; Fritsche, A.; et al. Circulating palmitoleate strongly and independently predicts insulin sensitivity in humans. Diabetes Care 2010, 33, 405–407. [Google Scholar] [CrossRef] [Green Version]
- Pinnick, K.E.; Neville, M.J.; Fielding, B.A.; Frayn, K.N.; Karpe, F.; Hodson, L. Gluteofemoral adipose tissue plays a major role in production of the lipokine palmitoleate in humans. Diabetes 2012, 61, 1399–1403. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Zeng, Y.; Xie, Y.; Huang, L.; Wu, Y. Bioinformatics analysis of the prognostic value of CCT6A and associated signalling pathways in breast cancer. Mol. Med. Rep. 2019, 19, 4344–4352. [Google Scholar] [CrossRef] [Green Version]
- Rome, S.; Clément, K.; Rabasa-Lhoret, R.; Loizon, E.; Poitou, C.; Barsh, G.S.; Riou, J.P.; Laville, M.; Vidal, H. Microarray profiling of human skeletal muscle reveals that insulin regulates approximately 800 genes during a hyperinsulinemic clamp. J. Biol. Chem. 2003, 278, 18063–18068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, B.; Wang, X.; Wu, Y.; Xu, C.; Xia, Z.; Dai, J.; Shao, M.; Zhao, F.; He, S.; Yang, L.; et al. The metabolic ER stress sensor IRE1α suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat. Immunol. 2017, 18, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Jäger, J.; Greiner, V.; Strzoda, D.; Seibert, O.; Niopek, K.; Sijmonsma, T.P.; Schäfer, M.; Jones, A.; De Guia, R.; Martignoni, M.; et al. Hepatic transforming growth factor-β 1 stimulated clone-22 D1 controls systemic cholesterol metabolism. Mol. Metab. 2014, 3, 155–166. [Google Scholar] [CrossRef]
- Akoumianakis, I.; Sanna, F.; Margaritis, M.; Badi, I.; Akawi, N.; Herdman, L.; Coutinho, P.; Fagan, H.; Antonopoulos, A.S.; Oikonomou, E.K.; et al. Adipose tissue-derived WNT5A regulates vascular redox signaling in obesity via USP17/RAC1-mediated activation of NADPH oxidases. Sci. Transl. Med. 2019, 11, eaav5055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Frühbeck, G.; Fernández-Quintana, B.; Paniagua, M.; Hernández-Pardos, A.W.; Valentí, V.; Moncada, R.; Catalán, V.; Becerril, S.; Gómez-Ambrosi, J.; Portincasa, P.; et al. FNDC4, a novel adipokine that reduces lipogenesis and promotes fat browning in human visceral adipocytes. Metabolism 2020, 108, 154261. [Google Scholar] [CrossRef] [PubMed]
- Leontovich, A.A.; Intine, R.V.; Sarras, M.P., Jr. Epigenetic Studies Point to DNA Replication/Repair Genes as a Basis for the Heritable Nature of Long Term Complications in Diabetes. J. Diabetes Res. 2016, 2016, 2860780. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Demographic Characteristics | Phenotypes and Cut-Offs (Mean ± SD) | ||
|---|---|---|---|
| N = 124 | Adipose Tissue (Dys)function | ||
| Sympton-Free Participants (Metabolic Risk Criteria) | ALR > 1 (N = 84) | ALR < 1 (N = 40) | p |
| ALR | 6.7 ± 10.0 | 0.6 ± 0.3 | <0.0001 |
| Age | 37 ± 14 | 39 ± 14 | 0.3887 |
| Weight (kg) | 70 ± 15 | 84 ± 16 | <0.0001 |
| Waist Circunference (cm) | 87 ± 12 | 99 ± 15 | <0.0001 |
| BMI (kg/m2) | 27 ± 5 | 32 ± 6 | <0.0001 |
| % Fat Total | 34 ± 9 | 40 ± 8 | <0.0001 |
| Systolic Pressure (mmHg) ≥ 130 | 111 ± 11 | 111 ± 14 | 0.7483 |
| Diastolic Pressure (mmHg) ≥ 85 | 70 ± 10 | 72 ± 10 | 0.3103 |
| Fasting Glucose (mg/dL) ≥ 100 | 97 ± 22 | 110 ± 29 | 0.0419 |
| 2-h Glucose (mg/dL) ≥ 140 | 127 ± 42 | 145 ± 54 | 0.0935 |
| Triglycerides (mg/dL) ≥ 150 | 118 ± 51 | 150 ± 57 | 0.0007 |
| HDL-cholesterol (mg/dL) < 40, Men; <50 Women | 48 ± 14 | 42 ± 11 | 0.0313 |
| High sensitive C-reactive protein (mg/L) (>90th percentile) * | 10.0 ± 13.8 | 19.6 ± 22.2 | 0.0027 |
| Whole body insulin resistance (Matsuda index) ** | 4.6 ± 4.0 | 2.7 ± 2.9 | <0.0001 |
| Immunometabolic and Postprandial Phenotypes | Phenotypes (Mean ± SD) | ||
|---|---|---|---|
| N = 124 | Adipose Tissue (Dys)function | ||
| Sympton-Free Participants | ALR > 1 (N = 84) | ALR < 1 (N = 40) | p |
| Adiponectin (μg/mL) | 32.7 ± 33.0 | 10.1 ± 5.2 | <0.0001 |
| Leptin (ng/mL) | 8.7 ± 6.5 | 21.0 ± 18.4 | <0.0001 |
| TNFa (pg/mL) | 5.0 ± 2.9 | 5.2 ± 2.8 | 0.6290 |
| IL-6 (pg/mL) | 9.1 ± 22.6 | 8.6 ± 9.5 | 0.0014 |
| MCP-1 pg/mL | 108.2 ± 143.9 | 107.1 ± 41.8 | 0.0544 |
| PAI-1 (pg/mL) | 34,169.0 ± 29,375.5 | 39,182.1 ± 30,956.3 | 0.2658 |
| Fibrinogen (mg/dL) | 107.7 ± 40.8 | 131.4 ± 73.4 | 0.0355 |
| Fasting Insulin (microU/mL) | 14.6 ± 18.6 | 25.2 ± 20.3 | <0.0001 |
| C-peptide (pg/mL) | 1.2 ± 0.5 | 1.9 ± 0.9 | <0.0001 |
| Insulin 120′ [microU/mL] | 64.8 ± 44.4 | 120.9 ± 74.4 | <0.0001 |
| HOMA-IR | 3.6 ± 4.7 | 7.2 ± 6.5 | <0.0001 |
| Glucagon (pg)mL) | 47.4 ± 64.2 | 46.5 ± 45.3 | 0.5168 |
| GLP-1 (pg/mL) | 13.0 ± 36.3 | 17.3 ± 44.3 | 0.8721 |
| Leptin AUC (5h) | 2598 | 6194 | 0.0002 |
| Insulin AUC (5h) | 16,847 | 28,158 | 0.0821 |
| Glucose AUC (5h) | 36,486 | 40,883 | 0.1124 |
| Triglycerides AUC 5h | 48,220 | 59,798 | 0.0025 |
| GLP-1 AUC 5h | 4743 | 5346 | 0.1306 |
| Glucagon AUC 5h | 18,487 | 17,435 | 0.1988 |
| C-peptide AUC 5h | 916.4 | 1246 | 0.0821 |
| Prevalence and Percentage of Individuals with Risk Phenotypes | |||
|---|---|---|---|
| Group 1 (N = 41) | Group 2 (N = 43) | Group 3 (N = 40) | |
| MH/MUH risk criteria and cut-offs | Mean ALR 9.5 ± 13.0 | Mean ALR 4.0 ± 4.5 | Mean ALR 0.6 ± 0.3 |
| Diabetic A1c > 6.5 | 0 (0.0%) | 3 (7.0%) | 4 (10.0%) |
| Prediabetic A1c 5.7–6.4 | 1 (2.4%) | 6 (14.0%) | 7 (17.5%) |
| Matsuda Index < 2.5 | 4 (9.8%) | 17 (39.5%) | 25 (62.5%) |
| HOMA-IR > 2.6 | 12 (29.3) | 16 (37.2%) | 28 (70.0%) |
| hsCRP > 35.7 | 0 (0.0%) | 4 (9.3%) | 8 (20.0%) |
| Glucose > 100 | 0 (0.0%) | 28 (65.1%) | 16 (40.0%) |
| Triglycerides > 150 | 3 (7.3%) | 17 (39.5%) | 16 (40.0%) |
| HDL < 40 Men < 50 Women | 16 (39.0%) | 36 (83.7%) | 33 (82.5%) |
| Dias BP > 85 | 1 (2.4%) | 5 (11.6%) | 4 (10.0%) |
| Sys BP > 130 | 0 (0.0%) | 5 (11.6%) | 4 (10.0%) |
| Waist > 88 Women - >102 Men | 3 (7.3%) | 26 (60.5%) | 30 (75.0%) |
| Demographic Characteristics | Mean Values for 14 Females | |||
|---|---|---|---|---|
| (H) ALR (N = 9) | SD (±) | (L) ALR (N = 5) | SD (±) | |
| Adipo/Lep Ratio | 2.2 | 1.1 | 0.5 | 0.4 |
| Age (Yr) | 38.5 | 11.8 | 32.6 | 13.0 |
| % Fat Total | 42.7 | 6.2 | 46.0 | 3.2 |
| Weight (kg) | 62.1 | 5.7 | 77.6 | 17.8 |
| Waist Circumference (cm) | 83.9 | 9.9 | 93.9 | 16.7 |
| BMI (kg/m2) | 26.8 | 3.1 | 32.8 | 7.8 |
| Triglycerides (mg/dL) | 119.8 | 42.2 | 145.0 | 60.1 |
| HDL-Cholesterol (mg/dL) | 41.4 | 9.6 | 47.8 | 9.4 |
| Adiponectin (μg/mL) | 24.9 | 15.5 | 7.0 | 2.8 |
| Leptin (ng/mL) | 11.1 | 3.9 | 21.0 | 11.3 |
| Fasting Glucose (mg/dL) | 87.0 | 6.9 | 86.2 | 8.4 |
| Glucose 120′ (mg/dL) | 121.2 | 21.8 | 107.0 | 7.0 |
| Fasting Insulin (microU/mL) | 7.1 | 4.2 | 17.3 | 9.5 |
| Insulin 120′ (microU/mL) | 59.5 | 42.4 | 101.7 | 42.2 |
| Matsuda Index | 6.6 | 4.1 | 2.9 | 1.3 |
| HOMA-IR | 1.5 | 0.9 | 3.7 | 2.0 |
| PAI-1 (pg/mL) | 42,867.5 | 31,186.8 | 56,408.4 | 57,940.0 |
| IL-6 (pg/mL) | 1.6 | 1.2 | 3.1 | 2.5 |
| TNFa (pg/mL) | 2.5 | 1.5 | 3.4 | 2.2 |
| MCP-1 pg/mL | 109.8 | 38.3 | 133.8 | 61.2 |
| Female ID (N = 6) | Gender | Age | % Fat | Adiponectin (μg/mL) | Leptin (ng/mL) | Adipo/Lep Ratio |
|---|---|---|---|---|---|---|
| MTY0003 | F | 49 | 34.5 | 18.37 | 7.41 | 2.48 |
| MTY0007 | F | 38 | 36.2 | 12.13 | 5.42 | 2.24 |
| MTY0006 | F | 33 | 50.3 | 23.73 | 11.50 | 2.06 |
| MTY0014 | F | 45 | 49.9 | 16.96 | 15.03 | 1.13 |
| Mean | 41 | 42.7 | 17.80 | 9.84 | 1.98 | |
| SD (±) | 7 | 8.5 | 4.77 | 4.29 | 0.59 | |
| MTY0009 | F | 20 | 43.2 | 8.46 | 8.93 | 0.95 |
| MTY0010 | F | 35 | 47 | 3.12 | 35.27 | 0.09 |
| Mean | 28 | 45.1 | 5.79 | 22.10 | 0.52 | |
| SD (±) | 10 | 2.7 | 3.78 | 18.62 | 0.61 |
| (H)ALR > 1 | (L)ALR < 1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Mean | SD (±) | SE | Mean | SD (±) | SE | Diff. | SE Diff. | p-Value |
| Waist Circumference (cm) | 82.5 | 7.5 | 4.33 | 90 | 18.08 | 10.44 | −7.5 | 11.3 | 0.543 |
| BMI (kg/m2) | 25.96 | 1.892 | 1.092 | 32.4 | 8.412 | 4.856 | −6.43 | 4.978 | 0.265 |
| % total fat | 40.33 | 8.673 | 5.007 | 46.7 | 3.36 | 1.939 | −6.36 | 5.37 | 0.301 |
| Fat Mass kg | 23.8 | 7.9 | 4.561 | 32.88 | 10.82 | 6.248 | −9.08 | 7.736 | 0.305 |
| Muscle Mass kg | 34.28 | 1.718 | 0.991 | 37.18 | 11.05 | 6.384 | −2.9 | 6.46 | 0.676 |
| Triglycerides (mg/dL) | 125.3 | 9.018 | 5.206 | 155.6 | 86.43 | 49.9 | −30.3 | 50.17 | 0.578 |
| Creatinin (mg/dL) | 0.6 | 0.264 | 0.152 | 0.666 | 0.152 | 0.088 | −0.06 | 0.176 | 0.724 |
| Uric acid (mg/dL) | 3.733 | 1.795 | 1.036 | 5.933 | 0.65 | 0.375 | −2.2 | 1.102 | 0.116 |
| BUN (mg/dL) | 8 | 4 | 2.309 | 9.333 | 0.577 | 0.333 | −1.33 | 2.333 | 0.598 |
| Total Cholesterol (mg/dL) | 162.3 | 80.22 | 46.31 | 156 | 11.13 | 6.429 | 6.333 | 46.76 | 0.898 |
| HDL (mg/dL) | 35 | 14.79 | 8.544 | 47.66 | 13.27 | 7.666 | −12.6 | 11.47 | 0.331 |
| LDL (mg/dL) | 110 | 59.02 | 34.07 | 79 | 9.539 | 5.507 | 31 | 34.52 | 0.419 |
| VLDL (mg/dL) | 17.33 | 6.429 | 3.711 | 29.33 | 17 | 9.82 | −12 | 10.49 | 0.316 |
| Alt (U/L) | 16.66 | 5.859 | 3.382 | 28.33 | 12.74 | 7.356 | −11.6 | 8.096 | 0.223 |
| Ast (U/L) | 33.33 | 12.89 | 7.446 | 55.33 | 18.82 | 10.86 | −22 | 13.17 | 0.17 |
| Alk phos (U/L) | 73.33 | 24.54 | 14.16 | 49.66 | 32.12 | 18.55 | 23.66 | 23.34 | 0.367 |
| Adipo/lep Ratio | 2.804 | 1.448 | 0.836 | 0.627 | 0.213 | 0.123 | 2.177 | 0.845 | 0.061 * |
| PAI-1 (pg/mL) | 21,095 | 28,057 | 16,198 | 68,896 | 46,810 | 27,026 | −4780 | 31,509 | 0.203 |
| MCP-1 (pg/mL) | 98.3 | 20.55 | 11.86 | 124.7 | 33.41 | 19.28 | −26.4 | 22.64 | 0.307 |
| IL-6 (pg/mL) | 0.656 | 0.705 | 0.407 | 2.833 | 0.667 | 0.385 | −2.17 | 0.56 | 0.017 ** |
| TNF-a (pg/mL) | 2.169 | 1.47 | 0.849 | 3.035 | 1.115 | 0.644 | −0.86 | 1.065 | 0.461 |
| hsCRP (mg/L) | 0.058 | 0.029 | 0.017 | 0.304 | 0.165 | 0.095 | −0.24 | 0.096 | 0.063 * |
| Matsuda Index | 8.991 | 5.405 | 3.12 | 3.313 | 1.667 | 0.962 | 5.678 | 3.265 | 0.157 |
| HOMA-IR | 0.905 | 0.393 | 0.226 | 3.39 | 2.88 | 1.663 | −2.48 | 1.678 | 0.212 |
| AUC Glucose | 350 | 61.55 | 35.53 | 310.9 | 2.155 | 1.244 | 39.08 | 35.56 | 0.333 |
| AUC Insulin | 134.8 | 61.86 | 35.71 | 282.8 | 87.22 | 50.36 | −148 | 61.74 | 0.074 * |
| AUC GLP-1 | 304.8 | 52.32 | 30.21 | 470.3 | 236.1 | 136.3 | −165 | 139.6 | 0.301 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallegos-Cabriales, E.C.; Rodriguez-Ayala, E.; Laviada-Molina, H.A.; Nava-Gonzalez, E.J.; Salinas-Osornio, R.A.; Orozco, L.; Leal-Berumen, I.; Castillo-Pineda, J.C.; Gonzalez-Lopez, L.; Escudero-Lourdes, C.; et al. Replication of Integrative Data Analysis for Adipose Tissue Dysfunction, Low-Grade Inflammation, Postprandial Responses and OMICs Signatures in Symptom-Free Adults. Biology 2021, 10, 1342. https://doi.org/10.3390/biology10121342
Gallegos-Cabriales EC, Rodriguez-Ayala E, Laviada-Molina HA, Nava-Gonzalez EJ, Salinas-Osornio RA, Orozco L, Leal-Berumen I, Castillo-Pineda JC, Gonzalez-Lopez L, Escudero-Lourdes C, et al. Replication of Integrative Data Analysis for Adipose Tissue Dysfunction, Low-Grade Inflammation, Postprandial Responses and OMICs Signatures in Symptom-Free Adults. Biology. 2021; 10(12):1342. https://doi.org/10.3390/biology10121342
Chicago/Turabian StyleGallegos-Cabriales, Esther C., Ernesto Rodriguez-Ayala, Hugo A. Laviada-Molina, Edna J. Nava-Gonzalez, Rocío A. Salinas-Osornio, Lorena Orozco, Irene Leal-Berumen, Juan Carlos Castillo-Pineda, Laura Gonzalez-Lopez, Claudia Escudero-Lourdes, and et al. 2021. "Replication of Integrative Data Analysis for Adipose Tissue Dysfunction, Low-Grade Inflammation, Postprandial Responses and OMICs Signatures in Symptom-Free Adults" Biology 10, no. 12: 1342. https://doi.org/10.3390/biology10121342