Synergistic Antibiotic Activity of Ricini Semen Extract with Oxacillin against Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Two MRSA Strains: SCCmec Type and Minimum Inhibitory Concentration
2.2. Analysis of the Chemical Composition of the Ricini Semen Extract by Gas Chromatography
2.3. Effects on the Cell Growth
2.4. Synergistic Activity of Oxacillin on MRSAs
2.5. Changes in Cellular Membrane Fluidity by Ricini Semen Extract and Its Component Fatty Acids
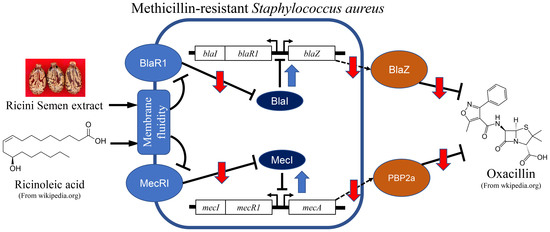
2.6. Changes in Antibiotic Resistance-Related Gene Expression by Ricini Semen Extract and Its Component Fatty Acids
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
3.2. Chemicals
3.3. Preparation of Ricini Semen Ethanol Extract
3.4. Identification of SCCmec Type of MRSA Strains
3.5. Measuring the Minimal Inhibition Concentration of the MRSA Strains
3.6. Analysis of the Chemical Composition of the Ricini Semen Extract by Gas Chromatography
3.7. Measuring the Growth Curve
3.8. Measuring Colony-Forming Units and Evaluating Synergistic Growth Inhibition
3.9. Gene Expression Analysis using Real-Time Polymerase Chain Reaction
3.10. Analysis of Bacterial Cellular Membrane Fluidity Using Laurdan
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CFU | Colony-forming units |
| MDR | Multidrug resistance |
| MIC | Minimum inhibitory concentration |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| PCR | Polymerase chain reaction |
| RT-PCR | Real-time polymerase chain reaction |
| SCCmec | Staphylococcal cassette chromosome mec |
| TSA | Tryptic Soy Agar |
| TSB | Tryptic Soy Broth |
References
- Hofer, U. The Cost of Antimicrobial Resistance. Nat. Rev. Microbiol. 2019, 17, 3. [Google Scholar] [CrossRef]
- Hassani, S.; Pakbin, B.; Brück, W.M.; Mahmoudi, R.; Mousavi, S. Prevalence, antibiotic resistance, toxin-typing and genotyping of Clostridium perfringens in raw beef meats obtained from Qazvin city, Iran. Antibiotics 2022, 11, 340. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Yoon, E.-J.; Kim, D.; Jeong, S.H.; Won, E.J.; Shin, J.H.; Kim, S.H.; Shin, J.H.; Shin, K.S.; Kim, Y.A. Antimicrobial Resistance of Major Clinical Pathogens in South Korea, May 2016 to April 2017: First one-year report from Kor-GLASS. Eurosurveillance 2018, 23, 1800047. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.A.; Irvine, A.D.; Foster, T.J. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends Microbiol. 2018, 26, 484–497. [Google Scholar] [CrossRef]
- Brauweiler, A.M.; Goleva, E.; Leung, D.Y. Staphylococcus aureus Lipoteichoic Acid Damages the Skin Barrier through an IL-1–Mediated Pathway. J. Investig. Dermatol. 2019, 139, 1753–1761.e4. [Google Scholar] [CrossRef]
- Okwu, M.U.; Olley, M.; Akpoka, A.O.; Izevbuwa, O.E. Methicillin-resistant Staphylococcus aureus (MRSA) and Anti-MRSA Activities of Extracts of Some Medicinal Plants: A Brief Review. AIMS Microbiol. 2019, 5, 117–137. [Google Scholar] [CrossRef]
- Michel, M.; Gutmann, L. Methicillin-resistant Staphylococus aureus and Vancomycin-Resistant Enterococci: Therapeutic Realities and Possibilities. Lancet 1997, 349, 1901–1906. [Google Scholar] [CrossRef]
- Tsai, H.-C.; Tao, C.-W.; Hsu, B.-M.; Yang, Y.-Y.; Tseng, Y.-C.; Huang, T.-Y.; Huang, S.-W.; Kuo, Y.-J.; Chen, J.-S. Multidrug-resistance in Methicillin-resistant Staphylococcus aureus (MRSA) Isolated from a Subtropical River Contaminated by nearby Livestock Industries. Ecotoxicol. Environ. Saf. 2020, 200, 110724. [Google Scholar] [CrossRef]
- Dugassa, J.; Shukuri, N. Review on Antibiotic Resistance and its Mechanism of Development. J. Health Med. Nurs. 2017, 1, 1–17. [Google Scholar]
- Niederman, M.S. Treatment Options for Nosocomial Pneumonia due to MRSA. J. Infect. 2009, 59, S25–S31. [Google Scholar] [CrossRef]
- Conly, J.; Johnston, B. Where are All the New Antibiotics? The New Antibiotic Paradox. Can. J. Infect. Dis. Med. Microbiol. 2005, 16, 159–160. [Google Scholar] [CrossRef]
- Spellberg, B.; Powers, J.H.; Brass, E.P.; Miller, L.G.; Edwards, J.E., Jr. Trends in Antimicrobial Drug Development: Implications for the Future. Clin. Infect. Dis. 2004, 38, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Wagner, H.; Ulrich-Merzenich, G. Synergy Research: Approaching a New Generation of Phytopharmaceuticals. Phytomedicine 2009, 16, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.H.; Houck, H.J. Synergy of Daptomycin with Oxacillin and Other β-Lactams against Methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 2871–2875. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Zhang, X.; Li, Z.; Yuan, J.; Shen, F.; Zhang, S. Synergistic Effect and Antibiofilm Activity of an Antimicrobial Peptide with Traditional Antibiotics against Multi-drug Resistant Bacteria. Microb. Pathog. 2021, 158, 105056. [Google Scholar] [CrossRef]
- Lainson, J.C.; Daly, S.M.; Triplett, K.; Johnston, S.A.; Hall, P.R.; Diehnelt, C.W. Synthetic Antibacterial Peptide Exhibits Synergy with Oxacillin against MRSA. ACS Med. Chem. Lett. 2017, 8, 853–857. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Hu, Z.-Q.; Hara, Y.; Shimamura, T. Inhibition of Penicillinase by Epigallocatechin Gallate Resulting in Restoration of Antibacterial Activity of Penicillin against Penicillinase-producing Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2266–2268. [Google Scholar] [CrossRef]
- Fujita, M.; Shiota, S.; Kuroda, T.; Hatano, T.; Yoshida, T.; Mizushima, T.; Tsuchiya, T. Remarkable Synergies between Baicalein and Tetracycline, and Baicalein and β-Lactams against Methicillin-Resistant Staphylococcus aureus. Microbiol. Immunol. 2005, 49, 391–396. [Google Scholar] [CrossRef]
- Scarpa, A.; Guerci, A. Various Uses of the Castor Oil Plant (Ricinus communis L.) a Review. J. Eethnopharmacol. 1982, 5, 117–137. [Google Scholar] [CrossRef]
- Kazeem, O.; Taiwo, O.; Kazeem, A.; Mondiu, D. Determination of Some Physical Properties of Castor (Ricirus communis) Oil. Int. J. Sci. Eng. Technol. 2014, 3, 1503–1508. [Google Scholar]
- Khan, I.A.; Abourashed, E.A. Leung’s Encyclopedia of Common Natural Ingredients: Used in Food, Drugs and Cosmetics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Naughton, F.C. Production, Chemistry, and Commercial Applications of Various Chemicals from Castor Oil. J. Am. Oil Chem. Soc. 1974, 51, 65–71. [Google Scholar] [CrossRef]
- Mohammed, M.A.Y.A. Antibacterial Activity of Raw Castor (Ricinus communis) Oil Against Escherichia coli and Staphylococcus aureus. Master Thesis, University of Gezira, Wad Medani, Sudan, 2021. [Google Scholar]
- Valera, M.C.; Maekawa, L.E.; Chung, A.; de Oliveira, L.D.; Carvalho, C.; Koga-Ito, C.Y.; Jorge, A. Effectiveness of Castor Oil Extract on Escherichia coli and its Endotoxins in Root Canals. Gen. Dent. 2012, 60, e204–e209. [Google Scholar] [PubMed]
- Kim, I.J.; Nam, S.Y.; Kim, M.J.; Rho, C.W.; Yun, T.; Kim, H.S.; Song, H.L.; Jeong, H.S. Analysis of Crude Fat and Fatty Acid in Collections of Ricinus communis L. Korean J. Med. Crop. Sci. 2008, 16, 301–305. [Google Scholar]
- Canoira, L.; Galeán, J.G.; Alcántara, R.; Lapuerta, M.; García-Contreras, R. Fatty Acid Methyl Esters (FAMEs) from Castor Oil: Production Process Assessment and Synergistic Effects in its Properties. Renew. Energy 2010, 35, 208–217. [Google Scholar] [CrossRef]
- Bonam, S.P. Preparation and Evaluation of Pluronic Lecithin Organogel Containing Ricinoleic Acid for Transdermal Drug Delivery. Master Thesis, University of Toledo, Toledo, OH, USA, 2013. [Google Scholar]
- Persson, C.; Robert, E.; Carlsson, E.; Robo, C.; López, A.; Godoy-Gallardo, M.; Ginebra, M.-P.; Engqvist, H. The Effect of Unsaturated Fatty Acid and Triglyceride Oil Addition on the Mechanical and Antibacterial Properties of Acrylic Bone Cements. J. Biomater. Appl. 2015, 30, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Sbihi, H.M.; Nehdi, I.A.; Mokbli, S.; Romdhani-Younes, M.; Al-Resayes, S.I. Hexane and Ethanol Extracted Seed Oils and Leaf Essential Compositions from Two Castor Plant (Ricinus communis L.) Varieties. Ind. Crop. Prod. 2018, 122, 174–181. [Google Scholar] [CrossRef]
- Song, H.-S.; Choi, T.-R.; Bhatia, S.K.; Lee, S.M.; Park, S.L.; Lee, H.S.; Kim, Y.-G.; Kim, J.-S.; Kim, W.; Yang, Y.-H. Combination Therapy using Low-concentration Oxacillin with Palmitic Acid and Span85 to Control Clinical Methicillin-resistant Staphylococcus aureus. Antibiotics 2020, 9, 682. [Google Scholar] [CrossRef] [PubMed]
- Greenway, D.; Dyke, K. Mechanism of the Inhibitory Action of Linoleic Acid on the Growth of Staphylococcus aureus. Microbiology 1979, 115, 233–245. [Google Scholar] [CrossRef]
- Kirby, W.M.; Rosenfeld, L.S.; Brodie, J. Oxacillin: Laboratory and Clinical Evaluation. JAMA 1962, 181, 739–744. [Google Scholar] [CrossRef]
- Sanford, J.P.; Reinarz, J.A. Complications Associated with Antibiotic Therapy. Dis. Mon. 1964, 10, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Berger-Bächi, B.; Rohrer, S. Factors Influencing Methicillin Resistance in Staphylococci. Arch. Microbiol. 2002, 178, 165–171. [Google Scholar] [CrossRef]
- Gong, R.; Lee, D.Y.; Lee, J.W.; Choi, D.J.; Kim, G.-S.; Lee, S.H.; Lee, Y.-S. Potentiating Activity of Rhein in Targeting of Resistance Genes in Methicillin-resistant Staphylococcus aureus. Asian Pac. J. Trop. Med. 2019, 12, 14–18. [Google Scholar] [CrossRef]
- Safo, M.K.; Zhao, Q.; Ko, T.-P.; Musayev, F.N.; Robinson, H.; Scarsdale, N.; Wang, A.H.-J.; Archer, G.L. Crystal Structures of the BlaI Repressor from Staphylococcus aureus and its Complex with DNA: Insights into Transcriptional Regulation of the bla and mec Operons. J. Bacteriol. 2005, 187, 1833–1844. [Google Scholar] [CrossRef]
- Hiramatsu, K. Molecular Evolution of MRSA. Microbiol. Immunol. 1995, 39, 531–543. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Katayama, Y.; Yuzawa, H.; Ito, T. Molecular Genetics of Methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 2002, 292, 67–74. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Kim, D.; Yoon, E.-J.; Hong, J.S.; Choi, M.H.; Kim, H.S.; Kim, Y.R.; Kim, Y.A.; Uh, Y.; Shin, K.S.; Shin, J.H. Major Bloodstream Infection-Causing Bacterial Pathogens and Their Antimicrobial Resistance in South Korea, 2017-2019: Phase I Report From Kor-GLASS. Front. Microbiol 2022, 12, 799084. [Google Scholar] [CrossRef]
- Wikler, M.A. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard. Clin. Lab. Stand. Inst. 2006, 26, M7-A7. [Google Scholar]
- Parsons, J.B.; Yao, J.; Frank, M.W.; Jackson, P.; Rock, C.O. Membrane Disruption by Antimicrobial Fatty Acids Releases Low-molecular-weight Proteins from Staphylococcus aureus. J. Bacteriol. 2012, 194, 5294–5304. [Google Scholar] [CrossRef]
- Kalhapure, R.S.; Mocktar, C.; Sikwal, D.R.; Sonawane, S.J.; Kathiravan, M.K.; Skelton, A.; Govender, T. Ion Pairing with Linoleic Acid Simultaneously Enhances Encapsulation Efficiency and Antibacterial Activity of Vancomycin in Solid Lipid Nanoparticles. Colloids Surf. B Biointerfaces 2014, 117, 303–311. [Google Scholar] [CrossRef]
- Bessa, L.J.; Palmeira, A.; Gomes, A.S.; Vasconcelos, V.; Sousa, E.; Pinto, M.; Costa, P.M.d. Synergistic Effects between Thioxanthones and Oxacillin against Methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2015, 21, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Buommino, E.; Vollaro, A.; Nocera, F.P.; Lembo, F.; DellaGreca, M.; Martino, L.D.; Catania, M.R. Synergistic Effect of Abietic Acid with Oxacillin against Methicillin-resistant Staphylococcus pseudintermedius. Antibiotics 2021, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-H.; Kang, O.-H.; Zhou, T.; Kong, R.; Lee, S.-J.; Kang, D.-H.; Jung, H.-I.; Lee, Y.-S.; Kwon, D.-Y. The Antimicrobial Activity of Hwangheuk-san and Synergy Effect with Oxacillin against Methicillin-resistant Staphylococcus aureus. Korea J. Herb. 2016, 31, 93–98. [Google Scholar] [CrossRef]
- An, J.; Zuo, G.; Hao, X.; Wang, G.; Li, Z. Antibacterial and Synergy of a Flavanonol Rhamnoside with Antibiotics against Clinical Isolates of Methicillin-resistant Staphylococcus aureus (MRSA). Phytomedicine 2011, 18, 990–993. [Google Scholar] [CrossRef]
- Desbois, A.P.; Smith, V.J. Antibacterial Free Fatty Acids: Activities, Mechanisms of Action and Biotechnological Potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef]
- Parasassi, T.; De Stasio, G.; d’Ubaldo, A.; Gratton, E. Phase Fluctuation in Phospholipid Membranes Revealed by Laurdan Fluorescence. Biophys. J. 1990, 57, 1179–1186. [Google Scholar] [CrossRef]
- Parasassi, T.; Conti, F.; Gratton, E. Time-resolved Fluorescence Emission Spectra of Laurdan in Phospholipid Vesicles by Multifrequency Phase and Modlat Modulation Fluorometry. Cell Mol. Biol. 1986, 32, 103–108. [Google Scholar]
- Bessa, L.J.; Ferreira, M.; Gameiro, P. Evaluation of Membrane Fluidity of Multidrug-resistant Isolates of Escherichia coli and Staphylococcus aureus in Presence and Absence of Antibiotics. J. Photochem. Photobiol. B Biol. 2018, 181, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, S.; Catteau, L.; Boukricha, L.; Quetin-Leclercq, J.; Mingeot-Leclercq, M.-P. Effect of Ursolic and Oleanolic Acids on Lipid Membranes: Studies on MRSA and Models of Membranes. Antibiotics 2021, 10, 1381. [Google Scholar] [CrossRef]
- Catteau, L.; Reichmann, N.T.; Olson, J.; Pinho, M.G.; Nizet, V.; Van Bambeke, F.; Quetin-Leclercq, J. Synergy between Ursolic and Oleanolic Acids from Vitellaria paradoxa leaf extract and β-Lactams against Methicillin-resistant Staphylococcus aureus: In Vitro and in Vivo Activity and Underlying Mechanisms. Molecules 2017, 22, 2245. [Google Scholar] [CrossRef]
- Peng, J.; Mishra, B.; Khader, R.; Felix, L.; Mylonakis, E. Novel Cecropin-4 Derived Peptides against Methicillin-resistant Staphylococcus aureus. Antibiotics 2021, 10, 36. [Google Scholar] [CrossRef]
- Poulsen, M.Ø.; Jacobsen, K.; Thorsing, M.; Kristensen, N.R.; Clasen, J.; Lillebæk, E.M.; Skov, M.N.; Kallipolitis, B.H.; Kolmos, H.J.; Klitgaard, J.K. Thioridazine Potentiates the Effect of a Beta-lactam Antibiotic against Staphylococcus aureus Independently of mecA Expression. Res. Microbiol. 2013, 164, 181–188. [Google Scholar] [CrossRef]
- Batabyal, B.; Kundu, G.K.; Biswas, S. Methicillin-resistant Staphylococcus aureus: A brief review. Int. Res. J. Biol. Sci. 2012, 1, 65–71. [Google Scholar]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements. Classification of Staphylococcal Cassette Chromosome mec (SCCmec): Guidelines for Reporting Novel SCCmec Elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef]
- McKinney, T.K.; Sharma, V.K.; Craig, W.A.; Archer, G.L. Transcription of the Gene Mediating Methicillin Resistance in Staphylococcus aureus (mecA) is Corepressed but not Coinduced by Cognate mecA and β-Lactamase Regulators. J. Bacteriol. 2001, 183, 6862–6868. [Google Scholar] [CrossRef] [Green Version]
- Klitgaard, J.K.; Skov, M.N.; Kallipolitis, B.H.; Kolmos, H.J. Reversal of Methicillin Resistance in Staphylococcus aureus by Thioridazine. J. Antimicrob. Chemother. 2008, 62, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Chae, S.-W.; Im, G.J.; Chung, J.-W.; Song, J.-J. Eugenol: A Phyto-compound Effective against Methicillin-resistant and Methicillin-sensitive Staphylococcus aureus Clinical Strain Biofilms. PLoS ONE 2015, 10, e0119564. [Google Scholar] [CrossRef] [PubMed]
- Ham, Y.; Kim, T.-J. Plant Extracts Inhibiting Biofilm Formation by Streptococcus mutans without Antibiotic Activity. J. Korean Wood Sci. Technol. 2018, 46, 692–702. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of Multiplex PCRs for Staphylococcal Cassette Chromosome mec Type Assignment: Rapid Identification System for mec, ccr, and Major Differences in Junkyard Regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef]
- Chusri, S.; Siriyong, T.; Na-Phatthalung, P.; Voravuthikunchai, S.P. Synergistic Effects of Ethnomedicinal Plants of Apocynaceae Family and Antibiotics against Clinical Isolates of Acinetobacter baumannii. Asian Pac. J. Trop. Med. 2014, 7, 456–461. [Google Scholar] [CrossRef]
- Chumak, N.; Mosiolek, M.; Schoft, V.K. Sample Preparation and Fractionation of Arabidopsis thaliana Sperm and Vegetative Cell Nuclei by FACS. Bio-Protocol 2015, 5, e1664. [Google Scholar] [CrossRef]
- Goldstein, F.; Perutka, J.; Cuirolo, A.; Plata, K.; Faccone, D.; Morris, J.; Sournia, A.; Kitzis, M.D.; Ly, A.; Archer, G. Identification and Phenotypic Characterization of a β-Lactam-dependent, Methicillin-resistant Staphylococcus aureus Strain. Antimicrob. Agents Chemother. 2007, 51, 2514–2522. [Google Scholar] [CrossRef]
- Bittar, F.; Ouchenane, Z.; Smati, F.; Raoult, D.; Rolain, J.-M. MALDI-TOF-MS for Rapid Detection of Staphylococcal Panton–Valentine Leukocidin. Int. J. Antimicrob. Agents 2009, 34, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, J.; Jeong, S.-I.; Jahng, K.Y.; Yu, K.-Y. Antimicrobial Effects and Resistant Regulation of Magnolol and Honokiol on Methicillin-resistant Staphylococcus aureus. BioMed Res. Int. 2015, 2015, 283630. [Google Scholar] [CrossRef]
- Kong, R.; Kang, O.-H.; Seo, Y.-S.; Mun, S.-H.; Zhou, T.; Shin, D.-W.; Kwon, D.-Y. The Inhibition Effect of Chlorpromazine against the β-Lactam Resistance of MRSA. Asian Pac. J. Trop. Med. 2016, 9, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Zou, G.; Hari, T.P.; Wilt, I.K.; Zhu, W.; Galle, N.; Faizi, H.A.; Hendricks, G.L.; Tori, K.; Pan, W. A Selective Membrane-targeting Repurposed Antibiotic with Activity against Persistent Methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2019, 116, 16529–16534. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Strain | MIC (mg/L) | ||||||
|---|---|---|---|---|---|---|---|
| Oxacillin | Ricini Semen Extract | Ricinoleic Acid | Linoleic Acid | Oleic Acid | Palmitic Acid | Stearic Acid | |
| CCARM 3807 | 1024 | >1024 | 256 | >512 | >512 | >256 | >256 |
| CCARM 3820 | 32 | 256 | 256 | >512 | >512 | >256 | >256 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Seo, Y.; Kim, S.-G.; Choi, Y.; Kim, H.J.; Kim, T.-J. Synergistic Antibiotic Activity of Ricini Semen Extract with Oxacillin against Methicillin-Resistant Staphylococcus aureus. Antibiotics 2023, 12, 340. https://doi.org/10.3390/antibiotics12020340
Kim M, Seo Y, Kim S-G, Choi Y, Kim HJ, Kim T-J. Synergistic Antibiotic Activity of Ricini Semen Extract with Oxacillin against Methicillin-Resistant Staphylococcus aureus. Antibiotics. 2023; 12(2):340. https://doi.org/10.3390/antibiotics12020340
Chicago/Turabian StyleKim, Minjun, Yena Seo, Seon-Gyeong Kim, Yedam Choi, Hyun Jung Kim, and Tae-Jong Kim. 2023. "Synergistic Antibiotic Activity of Ricini Semen Extract with Oxacillin against Methicillin-Resistant Staphylococcus aureus" Antibiotics 12, no. 2: 340. https://doi.org/10.3390/antibiotics12020340