Systematic In Silico Assessment of Antimicrobial Resistance Dissemination across the Global Plasmidome
Abstract
:1. Introduction
2. Results and Discussion
2.1. Independent Mobilization and Dissemination of Antimicrobial Resistance Genes
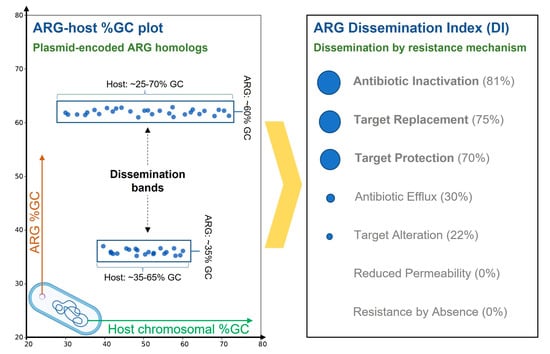
2.2. Resistance Mechanisms Determine the Prevalence of ARG Dissemination
2.3. Antimicrobial Agent Classes Present Different Dissemination Patterns
2.4. Conjugative Plasmids Mediate Dissemination of Antimicrobial Resistance Genes
3. Materials and Methods
3.1. Antimicrobial Resistance Gene Prediction, Classification and Plasmid Analysis
3.2. Antimicrobial Resistance Gene Dissemination Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, A.; Miller, W.R.; Arias, C.A. Mechanisms of Antimicrobial Resistance among Hospital-Associated Pathogens. Expert Rev. Anti-Infect. Ther. 2018, 16, 269–287. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (Ed.) Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2015.
- Berglund, B. Environmental Dissemination of Antibiotic Resistance Genes and Correlation to Anthropogenic Contamination with Antibiotics. Infect. Ecol. Epidemiol. 2015, 5, 28564. [Google Scholar] [CrossRef] [PubMed]
- Saima, S.; Fiaz, M.; Zafar, R.; Ahmed, I.; Arshad, M. Chapter 6—Dissemination of Antibiotic Resistance in the Environment. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Hashmi, M.Z., Ed.; Advances in Environmental Pollution Research series; Elsevier: Amsterdam, The Netherlands, 2020; Volume 1, pp. 99–116. ISBN 978-0-12-818882-8. [Google Scholar]
- Baym, M.; Lieberman, T.D.; Kelsic, E.D.; Chait, R.; Gross, R.; Yelin, I.; Kishony, R. Spatiotemporal Microbial Evolution on Antibiotic Landscapes. Science 2016, 353, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Suarez, S.A.; Martiny, A.C. Gene Amplification Uncovers Large Previously Unrecognized Cryptic Antibiotic Resistance Potential in E. Coli. Microbiol. Spectr. 2021, 9, e0028921. [Google Scholar] [CrossRef]
- van Hoek, A.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.; Aarts, H. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Mackie, R.I. Evolution and Ecology of Antibiotic Resistance Genes. FEMS Microbiol. Lett. 2007, 271, 147–161. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Domingues, S.; Nielsen, K.M. Membrane Vesicles and Horizontal Gene Transfer in Prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21. [Google Scholar] [CrossRef]
- Soler, N.; Forterre, P. Vesiduction: The Fourth Way of HGT. Environ. Microbiol. 2020, 22, 2457–2460. [Google Scholar] [CrossRef]
- Hernández-Beltrán, J.C.R.; San Millán, A.; Fuentes-Hernández, A.; Peña-Miller, R. Mathematical Models of Plasmid Population Dynamics. Front. Microbiol. 2021, 12, 606396. [Google Scholar] [CrossRef]
- Sánchez-Osuna, M.; Cortés, P.; Llagostera, M.; Barbé, J.; Erill, I. Exploration into the Origins and Mobilization of Di-Hydrofolate Reductase Genes and the Emergence of Clinical Resistance to Trimethoprim. Microb. Genom. 2020, 6, mgen000440. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Osuna, M.; Cortés, P.; Barbé, J.; Erill, I. Origin of the Mobile Di-Hydro-Pteroate Synthase Gene Determining Sulfonamide Resistance in Clinical Isolates. Front. Microbiol. 2019, 9, 3332. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of Antibiotic Resistance Genes from Antibiotic Producers to Pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef] [PubMed]
- Møller, T.S.B.; Liu, G.; Boysen, A.; Thomsen, L.E.; Lüthje, F.L.; Mortensen, S.; Møller-Jensen, J.; Olsen, J.E. Treatment with Cefotaxime Affects Expression of Conjugation Associated Proteins and Conjugation Transfer Frequency of an IncI1 Plasmid in Escherichia Coli. Front. Microbiol. 2017, 8, 2365. [Google Scholar] [CrossRef]
- Zeng, X.; Lin, J. Factors Influencing Horizontal Gene Transfer in the Intestine. Anim. Health Res. Rev. 2017, 18, 153–159. [Google Scholar] [CrossRef]
- Ellabaan, M.M.H.; Munck, C.; Porse, A.; Imamovic, L.; Sommer, M.O.A. Forecasting the Dissemination of Antibiotic Resistance Genes across Bacterial Genomes. Nat. Commun. 2021, 12, 2435. [Google Scholar] [CrossRef]
- Nielsen, T.K.; Browne, P.D.; Hansen, L.H. Antibiotic Resistance Genes Are Differentially Mobilized According to Resistance Mechanism. GigaScience 2022, 11, giac072. [Google Scholar] [CrossRef]
- Musto, H. How Many Factors Influence Genomic GC Content Among Prokaryotes? J. Mol. Evol. 2022, 91, 6–9. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Ochman, H. Amelioration of Bacterial Genomes: Rates of Change and Exchange. J. Mol. Evol. 1997, 44, 383–397. [Google Scholar] [CrossRef]
- Albornoz, E.; Tijet, N.; De Belder, D.; Gomez, S.; Martino, F.; Corso, A.; Melano, R.G.; Petroni, A. QnrE1, a Member of a New Family of Plasmid-Located Quinolone Resistance Genes, Originated from the Chromosome of Enterobacter Species. Antimicrob. Agents Chemother. 2017, 61, e02555-16. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R. Antibiotic Resistance Genes from the Environment: A Perspective through Newly Identified Antibiotic Resistance Mechanisms in the Clinical Setting. Clin. Microbiol. Infect. 2009, 15 (Suppl. S1), 20–25. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef]
- Walther-Rasmussen, J.; Høiby, N. OXA-Type Carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updates 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Hall, B.G. Determining the Limits of the Evolutionary Potential of an Antibiotic Resistance Gene. Mol. Biol. Evol. 2003, 20, 653–659. [Google Scholar] [CrossRef]
- Depardieu, F.; Podglajen, I.; Leclercq, R.; Collatz, E.; Courvalin, P. Modes and Modulations of Antibiotic Resistance Gene Expression. Clin. Microbiol. Rev. 2007, 20, 79–114. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What Is a Resistance Gene? Ranking Risk in Resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- Luo, X.; Yin, Z.; Zeng, L.; Hu, L.; Jiang, X.; Jing, Y.; Chen, F.; Wang, D.; Song, Y.; Yang, H.; et al. Chromosomal Integration of Huge and Complex Bla NDM-Carrying Genetic Elements in Enterobacteriaceae. Front. Cell. Infect. Microbiol. 2021, 11, 690799. [Google Scholar] [CrossRef]
- Shropshire, W.C.; Aitken, S.L.; Pifer, R.; Kim, J.; Bhatti, M.M.; Li, X.; Kalia, A.; Galloway-Peña, J.; Sahasrabhojane, P.; Arias, C.A.; et al. IS26-Mediated Amplification of BlaOXA-1 and BlaCTX-M-15 with Concurrent Outer Membrane Porin Disruption Associated with de Novo Carbapenem Resistance in a Recurrent Bacteraemia Cohort. J. Antimicrob. Chemother. 2021, 76, 385–395. [Google Scholar] [CrossRef]
- Aminov, R.I. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Shin, H.W.; Lim, J.; Kim, S.; Kim, J.; Kwon, G.C.; Koo, S.H. Characterization of Trimethoprim-Sulfamethoxazole Resistance Genes and Their Relatedness to Class 1 Integron and Insertion Sequence Common Region in Gram-Negative Bacilli. J. Microbiol. Biotechnol. 2015, 25, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-Resistant Staphylococcus Aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal Chromosomal Cassettes Mec (SCCmec): A Mobile Genetic Element in Methicillin-Resistant Staphylococcus Aureus. Microb. Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef] [PubMed]
- MacLean, R.C.; Hall, A.R.; Perron, G.G.; Buckling, A. The Population Genetics of Antibiotic Resistance: Integrating Molecular Mechanisms and Treatment Contexts. Nat. Rev. Genet. 2010, 11, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Slipski, C.J.; Jamieson-Datzkiw, T.R.; Zhanel, G.G.; Bay, D.C. Characterization of Proteobacterial Plasmid Integron-Encoded Qac Efflux Pump Sequence Diversity and Quaternary Ammonium Compound Antiseptic Selection in Escherichia Coli Grown Planktonically and as Biofilms. Antimicrob. Agents Chemother. 2021, 65, e0106921. [Google Scholar] [CrossRef]
- Nannini, E.; Murray, B.E.; Arias, C.A. Resistance or Decreased Susceptibility to Glycopeptides, Daptomycin, and Linezolid in Methicillin-Resistant Staphylococcus Aureus. Curr. Opin. Pharm. 2010, 10, 516–521. [Google Scholar] [CrossRef]
- Park, A.K.; Kim, H.; Jin, H.J. Phylogenetic Analysis of RRNA Methyltransferases, Erm and KsgA, as Related to Antibiotic Resistance. FEMS Microbiol. Lett. 2010, 309, 151–162. [Google Scholar] [CrossRef]
- Sharkey, L.K.R.; O’Neill, A.J. Antibiotic Resistance ABC-F Proteins: Bringing Target Protection into the Limelight. ACS Infect. Dis. 2018, 4, 239–246. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The Cfr RRNA Methyltransferase Confers Resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A Antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef]
- Ushtanit, A.; Kulagina, E.; Mikhailova, Y.; Makarova, M.; Safonova, S.; Zimenkov, D. Molecular Determinants of Ethionamide Resistance in Clinical Isolates of Mycobacterium Tuberculosis. Antibiotics 2022, 11, 133. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Ellington, M.J. The Emergence of Antibiotic Resistance by Mutation. Clin. Microbiol. Infect. 2007, 13, 5–18. [Google Scholar] [CrossRef]
- Baquero, F.; Alvarez-Ortega, C.; Martinez, J.L. Ecology and Evolution of Antibiotic Resistance. Environ. Microbiol. Rep. 2009, 1, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Alderliesten, J.B.; Duxbury, S.J.N.; Zwart, M.P.; de Visser, J.A.G.M.; Stegeman, A.; Fischer, E.A.J. Effect of Donor-Recipient Relatedness on the Plasmid Conjugation Frequency: A Meta-Analysis. BMC Microbiol. 2020, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.-C. Integron Involvement in Environmental Spread of Antibiotic Resistance. Front. Microbiol. 2012, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Bennett, P.M. Plasmid Encoded Antibiotic Resistance: Acquisition and Transfer of Antibiotic Resistance Genes in Bacteria. Br. J. Pharm. 2008, 153 (Suppl. S1), S347–S357. [Google Scholar] [CrossRef]
- Galata, V.; Fehlmann, T.; Backes, C.; Keller, A. PLSDB: A Resource of Complete Bacterial Plasmids. Nucleic Acids Res. 2019, 47, D195–D202. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG Database Update: Focus on Microbial Diversity, Model Organisms, and Widespread Pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Osuna, M.; Barbé, J.; Erill, I. Systematic In Silico Assessment of Antimicrobial Resistance Dissemination across the Global Plasmidome. Antibiotics 2023, 12, 281. https://doi.org/10.3390/antibiotics12020281
Sánchez-Osuna M, Barbé J, Erill I. Systematic In Silico Assessment of Antimicrobial Resistance Dissemination across the Global Plasmidome. Antibiotics. 2023; 12(2):281. https://doi.org/10.3390/antibiotics12020281
Chicago/Turabian StyleSánchez-Osuna, Miquel, Jordi Barbé, and Ivan Erill. 2023. "Systematic In Silico Assessment of Antimicrobial Resistance Dissemination across the Global Plasmidome" Antibiotics 12, no. 2: 281. https://doi.org/10.3390/antibiotics12020281