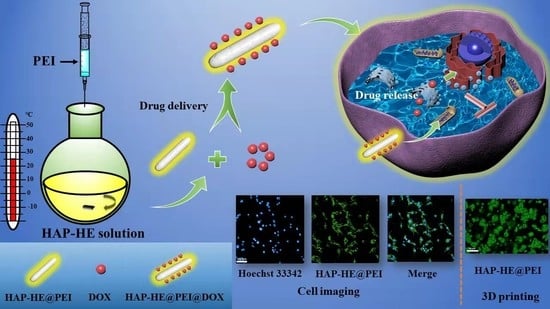
Surface-Fabrication of Fluorescent Hydroxyapatite for Cancer Cell Imaging and Bio-Printing Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Characterization
2.3. Preparation of HAP-HE
2.4. Preparation of HAP-HE@PEI
2.5. Encapsulation Efficiency and Release Characteristic of HAP-HE@PEI
2.6. Cytotoxicity Evaluation
2.7. High Content Cellular Imaging
2.8. HAP-HE@PEI for Cell Imaging of 3D Cultured Cells (Trial)
3. Results
3.1. Morphological Characteristics of HAP and HAP-HE@PEI
3.2. Crystal Structure Comparison of HAP, HAP-HE, HAP-HE@PEI
3.3. Element Contents and Changes of HAP, HAP-HE, HAP-HE@PEI
3.4. Fourier Transform Infrared Spectroscopy (FT-IR) Spectra of HAP, HAP-HE, HAP-HE@PEI
3.5. Thermogravimetric analysis of HAP, HAP-HE and HAP-HE@PEI
3.6. Fluorescence and UV Properties of HAP, HAP-HE and HAP-HE@PEI
3.7. Cytotoxicity Assay
3.8. The Release Characteristic of HAP-HE@PEI@DOX at pH 7.2 and pH 5.4
3.9. Cell Imaging of 2D Cultured Cells
3.10. Cell Imaging of 3D Cultured Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Guo, Q.; Chen, W.; Xie, W.; Wang, Y.; Wang, M.; You, T.; Pan, G. Biomimetic design of photonic materials for biomedical applications. Acta Biomater. 2021, 121, 143–179. [Google Scholar] [CrossRef] [PubMed]
- Broadwater, D.; Medeiros, H.C.; Lunt, R.R.; Lunt, S.Y. Current Advances in Photoactive Agents for Cancer Imaging and Therapy. Annu. Rev. Biomed. Eng. 2021, 23, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Yodsanit, N.; Wang, B.; Zhao, Y.; Guo, L.W.; Kent, K.C.; Gong, S. Recent progress on nanoparticles for targeted aneurysm treatment and imaging. Biomaterials 2021, 265, 120406. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Zhang, J.; Zhai, J.; Hong, J.; Yuan, C.; Liang, M. Ferritin: A Multifunctional Nanoplatform for Biological Detection, Imaging Diagnosis, and Drug Delivery. Acc. Chem. Res. 2021, 54, 3313–3325. [Google Scholar] [CrossRef] [PubMed]
- Wong, X.Y.; Sena-Torralba, A.; Alvarez-Diduk, R.; Muthoosamy, K.; Merkoçi, A. Nanomaterials for Nanotheranostics: Tuning Their Properties According to Disease Needs. ACS Nano 2020, 14, 2585–2627. [Google Scholar] [CrossRef]
- Akhlaghi, S.P.; Berry, R.C.; Tam, K.C.J.C. Nanocelluloses as a Novel Vehicle for Controlled Drug Delivery. In Handbook of Nanocelluloses; Springer International Publishing: Cham, Switzerland, 2013; pp. 1747–1764. [Google Scholar]
- Bazaka, K.; Jacob, M.V.; Crawford, R.J.; Ivanova, E.P. Plasma-assisted surface modification of organic biopolymers to prevent bacterial attachment. Acta Biomater. 2011, 7, 2015–2028. [Google Scholar] [CrossRef]
- Ebrahimi, S.B.; Samanta, D.; Mirkin, C.A. DNA-Based Nanostructures for Live-Cell Analysis. J. Am. Chem. Soc. 2020, 142, 11343–11356. [Google Scholar] [CrossRef]
- Rauschenbach, M.; Lawrenson, S.B.; Taresco, V.; Pearce, A.K.; O’Reilly, R.K. Antimicrobial Hyperbranched Polymer-Usnic Acid Complexes through a Combined ROP-RAFT Strategy. Macromol. Rapid Commun. 2020, 41, e2000190. [Google Scholar] [CrossRef]
- Christopherson, C.J.; Paisley, N.R.; Xiao, Z.; Algar, W.R.; Hudson, Z.M. Red-Emissive Cell-Penetrating Polymer Dots Exhibiting Thermally Activated Delayed Fluorescence for Cellular Imaging. J. Am. Chem. Soc. 2021, 143, 13342–13349. [Google Scholar] [CrossRef]
- Wang, Y.P.; Wu, K.; Pan, M.; Li, K.; Mo, J.T.; Duan, X.H.; He, H.Z.; Shen, J.; Su, C.Y. One-/Two-Photon Excited Cell Membrane Imaging and Tracking by a Photoactive Nanocage. ACS Appl. Mater. Interfaces 2020, 12, 35873–35881. [Google Scholar] [CrossRef]
- Ji, X.; Yuan, X.; Ma, L.; Bi, B.; Zhu, H.; Lei, Z.; Liu, W.; Pu, H.; Jiang, J.; Jiang, X.; et al. Mesenchymal stem cell-loaded thermosensitive hydroxypropyl chitin hydrogel combined with a three-dimensional-printed poly(ε-caprolactone)/nano-hydroxyapatite scaffold to repair bone defects via osteogenesis, angiogenesis and immunomodulation. Theranostics 2020, 10, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Li, Y.; Zhang, A.; Zhang, H. Nano-hydroxyapatite incorporated gelatin/zein nanofibrous membranes: Fabrication, characterization and copper adsorption. Int. J. Biol. Macromol. 2020, 154, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Lin, W.; Zou, Y.; Xu, B.; Zhang, X.; Zhao, J.; Rong, J. Nano-hydroxyapatite enhanced double network hydrogels with excellent mechanical properties for potential application in cartilage repair. Carbohydr. Polym. 2020, 229, 115523. [Google Scholar] [CrossRef] [PubMed]
- Lara-Ochoa, S.; Ortega-Lara, W.; Guerrero-Beltrán, C.E. Hydroxyapatite Nanoparticles in Drug Delivery: Physicochemistry and Applications. Pharmaceutics 2021, 13, 1642. [Google Scholar] [CrossRef]
- El-Maghrabi, H.H.; Younes, A.A.; Salem, A.R.; Rabie, K.; El-Shereafy, E.S. Magnetically modified hydroxyapatite nanoparticles for the removal of uranium (VI): Preparation, characterization and adsorption optimization. J. Hazard. Mater. 2019, 378, 120703. [Google Scholar] [CrossRef]
- Li, D.; Huang, X.; Wu, Y.; Li, J.; Cheng, W.; He, J.; Tian, H.; Huang, Y. Preparation of pH-responsive mesoporous hydroxyapatite nanoparticles for intracellular controlled release of an anticancer drug. Biomater. Sci. 2016, 4, 272–280. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, S.; Qiu, J.; Li, J.; Sang, Y.; Xia, H.; Jiang, H.; Claverie, J.; Liu, H. Eu/Tb codoped spindle-shaped fluorinated hydroxyapatite nanoparticles for dual-color cell imaging. Nanoscale 2016, 8, 11580–11587. [Google Scholar] [CrossRef]
- He, Y.; Lv, C.; Hou, X.; Wu, L. Mono-dispersed nano-hydroxyapatite based MRI probe with tetrahedral DNA nanostructures modification for in vitro tumor cell imaging. Anal. Chim. Acta 2020, 1138, 141–149. [Google Scholar] [CrossRef]
- Hui, J.; Zhang, X.; Zhang, Z.; Wang, S.; Tao, L.; Wei, Y.; Wang, X. Fluoridated HAp: Ln3+ (Ln = Eu or Tb) nanoparticles for cell-imaging. Nanoscale 2012, 4, 6967–6970. [Google Scholar] [CrossRef]
- Zhou, R.; Li, M.; Wang, S.; Wu, P.; Wu, L.; Hou, X. Low-toxic Mn-doped ZnSe@ZnS quantum dots conjugated with nano-hydroxyapatite for cell imaging. Nanoscale 2014, 6, 14319–14325. [Google Scholar] [CrossRef]
- Heng, C.; Zheng, X.; Liu, M.; Xu, D.; Huang, H.; Deng, F.; Hui, J.; Zhang, X.; Wei, Y. Fabrication of Luminescent Hydroxyapatite Nanorods through Surface-initiated RAFT. Appl. Surf. Sci. 2016, 386, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Steeg, P.S. Endocytosis: A pivotal pathway for regulating metastasis. Br. J. Cancer 2021, 124, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Guo, H.; Edman, M.; Hamm-Alvarez, S.F. Application of advances in endocytosis and membrane trafficking to drug delivery. Adv. Drug Deliv. Rev. 2020, 157, 118–141. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, F.; Liu, X.; Sang, Y.; Zhang, L.; Ren, J.; Qu, X. Cell membrane-camouflaged liposomes for tumor cell-selective glycans engineering and imaging in vivo. Proc. Natl. Acad. Sci. USA 2021, 118, e2022769118. [Google Scholar] [CrossRef]
- Fontana, F.; Lindstedt, H.; Correia, A.; Chiaro, J.; Kari, O.K.; Ndika, J.; Alenius, H.; Buck, J.; Sieber, S.; Mäkilä, E.; et al. Influence of Cell Membrane Wrapping on the Cell-Porous Silicon Nanoparticle Interactions. Adv. Healthc. Mater. 2020, 9, e2000529. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Z.; Ruan, B.; Zhang, X.; Jiang, T.; Ma, N.; Tsai, F.C. Design and Synthesis of Polyimide Covalent Organic Frameworks. Macromol. Rapid Commun. 2020, 41, e2000402. [Google Scholar] [CrossRef]
- Alphandéry, E. Light-Interacting iron-based nanomaterials for localized cancer detection and treatment. Acta Biomater. 2021, 124, 50–71. [Google Scholar] [CrossRef]
- Kurian, A.G.; Singh, R.K.; Patel, K.D.; Lee, J.H.; Kim, H.W. Multifunctional GelMA platforms with nanomaterials for advanced tissue therapeutics. Bioact. Mater. 2021, 8, 267–295. [Google Scholar] [CrossRef]
- Hwangbo, H.; Lee, H.; Jin, E.J.; Lee, J.; Jo, Y.; Ryu, D.; Kim, G. Bio-printing of aligned GelMa-based cell-laden structure for muscle tissue regeneration. Bioact. Mater. 2021, 8, 57–70. [Google Scholar] [CrossRef]
- Santos, O.; Cancino-Bernardi, J.; Pincela Lins, P.M.; Sampaio, D.; Pavan, T.; Zucolotto, V. Near-Infrared Photoactive Theragnostic Gold Nanoflowers for Photoacoustic Imaging and Hyperthermia. ACS Appl. Bio Mater. 2021, 4, 6780–6790. [Google Scholar] [CrossRef]
- Biedrzycka, A.; Skwarek, E.; Hanna, U.M. Hydroxyapatite with magnetic core: Synthesis methods, properties, adsorption and medical applications. Adv. Colloid. Interface Sci. 2021, 291, 102401. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Pandey, S.K.; Arunan, E.; Srivastava, C. A review on hydroxyapatite coatings for the biomedical applications: Experimental and theoretical perspectives. J. Mater. Chem. B 2021, 9, 228–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; He, J.; Manandhar, P.; Yang, Y.; Liu, P.; Gu, N. Gauging surface charge distribution of live cell membrane by ionic current change using scanning ion conductance microscopy. Nanoscale 2021, 13, 19973–19984. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, P.; Cervera, J.; Ali, M.; Nasir, S.; Ensinger, W.; Mafe, S. Impact of Surface Charge Directionality on Membrane Potential in Multi-ionic Systems. J. Phys. Chem. Lett. 2020, 11, 2530–2534. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.L.; Nudelman, F.; Chu, C.H.; Lo, E.C.; Mei, M.L. The effects of strontium-doped bioactive glass and fluoride on hydroxyapatite crystallization. J. Dent. 2021, 105, 103581. [Google Scholar] [CrossRef]
- Ullah, M.; Kodam, S.P.; Mu, Q.; Akbar, A. Microbubbles versus extracellular vesicles as therapeutic cargo for targeting drug delivery. ACS Nano 2021, 15, 3612–3620. [Google Scholar] [CrossRef]
- Nichol, J.W.; Koshy, S.T.; Bae, H.; Hwang, C.M.; Yamanlar, S.; Khademhosseini, A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 2010, 31, 5536–5544. [Google Scholar] [CrossRef] [Green Version]
- Doctor, A.; Seifert, V.; Ullrich, M.; Hauser, S.; Pietzsch, J. Three-Dimensional Cell Culture Systems in Radiopharmaceutical Cancer Research. Cancers 2020, 12, 2765. [Google Scholar] [CrossRef]
- Dimitriou, R.; Jones, E.; McGonagle, D.; Giannoudis, P.V. Bone regeneration: Current concepts and future directions. BMC Med. 2011, 9, 66. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Zhang, H.; Wang, X. Microfluidic-Chip-Integrated Biosensors for Lung Disease Models. Biosensors 2021, 11, 456. [Google Scholar] [CrossRef] [PubMed]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, W.; Li, Z.; Wang, X.; Tian, F.; Yang, J. Surface-Fabrication of Fluorescent Hydroxyapatite for Cancer Cell Imaging and Bio-Printing Applications. Biosensors 2022, 12, 419. https://doi.org/10.3390/bios12060419
Wan W, Li Z, Wang X, Tian F, Yang J. Surface-Fabrication of Fluorescent Hydroxyapatite for Cancer Cell Imaging and Bio-Printing Applications. Biosensors. 2022; 12(6):419. https://doi.org/10.3390/bios12060419
Chicago/Turabian StyleWan, Weimin, Ziqi Li, Xi Wang, Fei Tian, and Jian Yang. 2022. "Surface-Fabrication of Fluorescent Hydroxyapatite for Cancer Cell Imaging and Bio-Printing Applications" Biosensors 12, no. 6: 419. https://doi.org/10.3390/bios12060419