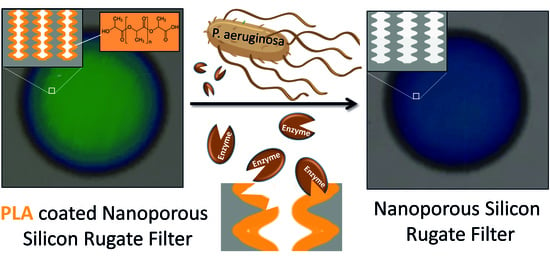
Bare Eye Detection of Bacterial Enzymes of Pseudomonas aeruginosa with Polymer Modified Nanoporous Silicon Rugate Filters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fabrication of pSiRF
2.2. Modification of pSiRF with PLA
2.3. Enzymatic Degradation of PLA with Proteinase K
2.4. Bacteria Tests
2.5. UV−Visible Spectroscopy
2.6. Reflectometric Interferometry Spectroscopy (RIfS)
2.7. Field Emission Scanning Electron Microscopy (FESEM)
3. Results
3.1. Fabrication of pSiRF Sensors
3.2. Stabilization of pSiRF Sensors
3.3. Modification of pSiRF with PLA
3.4. Enzymatic Degradation of PLA by Proteinase K
3.5. Enzymatic Degradation of PLA by P. aeruginosa
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic Resistance Increases with Local Temperature. Nat. Clim. Change 2018, 8, 510–514. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States. CDC Office of Infectious Diseases (OID). 2013. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 30 December 2020).
- Moore, J.L.; Caprioli, R.M.; Skaar, E.P. Advanced Mass Spectrometry Technologies for the Study of Microbial Pathogenesis. Curr. Opin. Microbiol. 2014, 19, 45–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vindenes, H.; Bjerknes, R. Microbial colonization of large wounds. Burns 1995, 21, 575–579. [Google Scholar] [CrossRef]
- Park, K.S.; Huang, C.-H.; Lee, K.; Yoo, Y.-E.; Castro, C.M.; Weissleder, R.; Lee, H. Rapid Identification of Health Care Associated Infections with an Integrated Fluorescence Anisotropy System. Sci. Adv. 2016, 2, e1600300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Multistate Point-Prevalence Survey of Health Care—Associated Infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef] [Green Version]
- Al-Zawity, J.; Afzal, F.; Awan, A.; Nordhoff, D.; Kleimann, A.; Wesner, D.; Montier, T.; Le Gall, T.; Müller, M. Effects of the sex steroid hormone estradiol on biofilm growth of cystic fibrosis Pseudomonas aeruginosa isolates. Front. Cell. Infect. Microbiol. 2022, 12, 941014. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-C.; Chu, C.-C.; Cheng, A.; Huang, Y.-T.; Hsueh, P.-R. Correlation Between Antimicrobial Consumption and Incidence of Health-Care-Associated Infections due to Methicillin-Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococci at a University Hospital in Taiwan from 2000 to 2010. J. Microbiol. Immunol. Infect. 2015, 48, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Figuero, E.; Sánchez-Beltrán, M.; Cuesta-Frechoso, S.; Tejerina, J.M.; del Castro, J.A.; Gutiérrez, J.M.; Herrera, D.; Sanz, M. Detection of Periodontal Bacteria in Atheromatous Plaque by Nested Polymerase Chain Reaction. J. Periodontol. 2011, 82, 1469–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodaka, H.; Iwata, M.; Yumoto, S.; Kashitani, F. Evaluation of a New Agar Medium Containing Cetrimide, Kanamycin and Nalidixic Acid for Isolation and Enhancement of Pigment Production of Pseudomonas aeruginosa in Clinical Samples. J. Basic Microbiol. 2003, 43, 407–413. [Google Scholar] [CrossRef]
- Baud, D.; Zufferey, J.; Hohlfeld, P.; Greub, G. Performance of an Automated Multiplex Immunofluorescence Assay for Detection of Chlamydia Trachomatis Tmmunoglobulin G. Diagn. Microbiol. Infect. Dis. 2014, 78, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Laabei, M.; Jamieson, W.D.; Lewis, S.E.; Diggle, S.P.; Jenkins, A.T.A. A New Assay for Rhamnolipid Detection-Important Virulence Factors of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2014, 98, 7199–7209. [Google Scholar] [CrossRef] [PubMed]
- Tücking, K.-S.; Grützner, V.; Unger, R.E.; Schönherr, H. Dual Enzyme-Responsive Capsules of Hyaluronic Acid-block-Poly(Lactic Acid) for Sensing Bacterial Enzymes. Macromol. Rapid Commun. 2015, 36, 1248–1254. [Google Scholar] [CrossRef]
- Thet, N.T.; Alves, D.R.; Bean, J.E.; Booth, S.; Nzakizwanayo, J.; Young, A.E.R.; Jones, B.V.; Jenkins, A.T.A. Prototype Development of the Intelligent Hydrogel Wound Dressing and Its Efficacy in the Detection of Model Pathogenic Wound Biofilms. ACS Appl. Mater. Interfaces 2016, 8, 14909–14919. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, M.-M.S.; Laabei, M.; Jenkins, A.T.A.; Schönherr, H. Autonomously Sensing Hydrogels for the Rapid and Selective Detection of Pathogenic Bacteria. Macromol. Rapid Commun. 2015, 36, 2123–2128. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Gwynne, L.; Sedgwick, A.C.; Müller, M.; Williams, G.T.; Jenkins, A.T.A.; James, T.D.; Schönherr, H. Enhanced Colorimetric Differentiation between Staphylococcus aureus and Pseudomonas aeruginosa Using a Shape-Encoded Sensor Hydrogel. ACS Appl. Bio Mater. 2020, 3, 4398–4407. [Google Scholar] [CrossRef] [PubMed]
- Thet, N.T.; Mercer-Chalmers, J.; Greenwood, R.J.; Young, A.E.R.; Coy, K.; Booth, S.; Sack, A.; Jenkins, A.T.A. SPaCE Swab: Point-of-Care Sensor for Simple and Rapid Detection of Acute Wound Infection. ACS Sens. 2020, 5, 2652–2657. [Google Scholar] [CrossRef] [PubMed]
- Krismastuti, F.S.H.; Bayat, H.; Voelcker, N.H.; Schönherr, H. Real Time Monitoring of Layer-by-Layer Polyelectrolyte Deposition and Bacterial Enzyme Detection in Nanoporous Anodized Aluminum Oxide. Anal. Chem. 2015, 87, 3856–3863. [Google Scholar] [CrossRef]
- Low, S.P.; Voelcker, N.H.; Canham, L.T.; Williams, K.A. The Biocompatibility of Porous Silicon in Tissues of the Eye. Biomaterials 2009, 30, 2873–2880. [Google Scholar] [CrossRef] [PubMed]
- Secret, E.; Smith, K.; Dubljevic, V.; Moore, E.; Macardle, P.; Delalat, B.; Rogers, M.-L.; Johns, T.G.; Durand, J.-O.; Cunin, F.; et al. Antibody-Functionalized Porous Silicon Nanoparticles for Vectorization of Hydrophobic Drugs. Adv. Healthc. Mater. 2013, 2, 718–727. [Google Scholar] [CrossRef]
- Janshoff, A.; Dancil, K.-P.S.; Steinem, C.; Greiner, D.P.; Lin, V.S.-Y.; Gurtner, C.; Motesharei, K.; Sailor, M.J.; Ghadiri, M.R. Macroporous p-Type Silicon Fabry−Perot Layers. Fabrication, Characterization, and Applications in Biosensing. J. Am. Chem. Society. 1998, 120, 12108–12116. [Google Scholar] [CrossRef]
- Müller, S.; Cavallaro, A.; Vasilev, K.; Voelcker, N.H.; Schönherr, H. Temperature-Controlled Antimicrobial Release from Poly(diethylene glycol methylether methacrylate)-Functionalized Bottleneck-Structured Porous Silicon for the Inhibition of Bacterial Growth. Macromol. Chem. Phys. 2016, 217, 2243–2251. [Google Scholar] [CrossRef]
- Rendina, I.; Rea, I.; Rotiroti, L.; de Stefano, L. Porous Silicon-Based Optical Biosensors and Biochips. Phys. E Low-Dimens. Syst. Nanostructures 2007, 38, 188–192. [Google Scholar] [CrossRef]
- Piya, R.; Gupta, B.; Gooding, J.J.; Reece, P.J. Optimising Porous Silicon Bragg Reflectors for Narrow Spectral Resonances. J. Appl. Phys. 2018, 124, 163103. [Google Scholar] [CrossRef]
- Szili, E.J.; Jane, A.; Low, S.P.; Sweetman, M.; Macardle, P.; Kumar, S.; Smart, R.C.S.; Voelcker, N.H. Interferometric Porous Silicon Transducers Using an Enzymatically Amplified Optical Signal. Sens. Actuators B Chem. 2011, 160, 341–348. [Google Scholar] [CrossRef]
- Stefano, L.D.; Rotiroti, L.; Rea, I.; Moretti, L.; Francia, G.D.; Massera, E.; Lamberti, A.; Arcari, P.P.; Sanges, C.; Rendina, I. Porous Silicon-Based Optical Biochips. J. Opt. A Pure Appl. Opt. 2006, 8, S540–S544. [Google Scholar] [CrossRef]
- Kilian, K.A.; Lai, L.M.H.; Magenau, A.; Cartland, S.; Böcking, T.; Di Girolamo, N.; Gal, M.; Gaus, K.; Gooding, J.J. Smart Tissue Culture: In Situ Monitoring of the Activity of Protease Enzymes Secreted from Live Cells using Nanostructured Photonic Crystals. Nano Lett. 2009, 9, 2021–2025. [Google Scholar] [CrossRef] [PubMed]
- Massad-Ivanir, N.; Shtenberg, G.; Tzur, A.; Krepker, M.A.; Segal, E. Engineering Nanostructured Porous SiO2 Surfaces for Bacteria Detection via “Direct Cell Capture”. Anal. Chem. 2011, 83, 3282–3289. [Google Scholar] [CrossRef]
- Tong, W.Y.; Sweetman, M.J.; Marzouk, E.R.; Fraser, C.; Kuchel, T.; Voelcker, N.H. Towards a Subcutaneous Optical Biosensor Based on Thermally Hydrocarbonised Porous Silicon. Biomaterials 2016, 74, 217–230. [Google Scholar] [CrossRef]
- Anderson, M.A.; Tinsley-Bown, A.; Allcock, P.; Perkins, E.A.; Snow, P.; Hollings, M.; Smith, R.G.; Reeves, C.; Squirrell, D.J.; Nicklin, S.; et al. Sensitivity of the Optical Properties of Porous Silicon Layers to the Refractive Index of Liquid in the Pores. Phys. Status Solidi 2003, 197, 528–533. [Google Scholar] [CrossRef]
- Bai, L.; Gao, Y.; Wang, J.; Aili, T.; Jia, Z.; Lv, X.; Huang, X.; Yang, J. Detection of Lactoglobulin by a Porous Silicon Microcavity Biosensor Based on the Angle Spectrum. Sensors 2022, 22, 1912. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, M.; Wang, X.; Wang, J.; Jia, Z.; Lv, X.; Huang, X. Spectral-Free Double Light Detection of DNA Based on a Porous Silicon Bragg Mirror. Sensors 2022, 22, 7048. [Google Scholar] [CrossRef] [PubMed]
- Orosco, M.M.; Pacholski, C.; Miskelly, G.M.; Sailor, M.J. Protein-Coated Porous-Silicon Photonic Crystals for Amplified Optical Detection of Protease Activity. Adv. Mater. 2006, 18, 1393–1396. [Google Scholar] [CrossRef]
- Tücking, K.-S.; Vasani, R.B.; Cavallaro, A.A.; Voelcker, N.H.; Schönherr, H.; Prieto-Simon, B. Hyaluronic Acid-Modified Porous Silicon Films for the Electrochemical Sensing of Bacterial Hyaluronidase. Macromol. Rapid Commun. 2018, 39, e1800178. [Google Scholar] [CrossRef] [PubMed]
- Sailor, M.J. Porous Silicon in Practice: Preparation, Characterization and Applications; Wiley-VCH: Weinheim, Germany, 2012. [Google Scholar]
- Reece, P.J.; Gal, M.; Tan, H.H.; Jagadish, C. Optical Properties of Erbium-Implanted Porous Silicon Microcavities. Appl. Phys. Lett. 2004, 85, 3363–3365. [Google Scholar] [CrossRef]
- Chhasatia, R.; Sweetman, M.J.; Prieto-Simon, B.; Voelcker, N.H. Performance Optimisation of Porous Silicon Rugate Filter Biosensor for the Detection of Insulin. Sens. Actuators B Chem. 2018, 273, 1313–1322. [Google Scholar] [CrossRef]
- Gupta, B.; Guan, B.; Reece, P.J.; Gooding, J.J. Porous Silicon Photonic Crystals for Detection of Infections. Proc. SPIE Biosensing Nanomed. V 2012, 8460, 136–143. [Google Scholar] [CrossRef]
- Shimpi, N.; Borane, M.; Mishra, S.; Kadam, M. Biodegradation of Polystyrene (PS)-Poly(Lactic Acid) (PLA) Nanocomposites Using Pseudomonas aeruginosa. Macromol. Res. 2012, 20, 181–187. [Google Scholar] [CrossRef]
- Ramot, Y.; Haim-Zada, M.; Domb, A.J.; Nyska, A. Biocompatibility and Safety of PLA and its Copolymers. Adv. Drug Deliv. Rev. 2016, 107, 153–162. [Google Scholar] [CrossRef]
- Oda, Y.; Yonetsu, A.; Urakami, T.; Tonomura, K. Degradation of Polylactide by Commercial Proteases. J. Polym. Environ. 2000, 8, 29–32. [Google Scholar] [CrossRef]
- Holloway, B.W. Genetic Recombination in Pseudomonas aeruginosa. Microbiology 1955, 13, 572–581. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orosco, M.M.; Pacholski, C.; Sailor, M.J. Real-Time Monitoring of Enzyme Activity in a Mesoporous Silicon Double Layer. Nat. Nanotechnol. 2009, 4, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Pap, A.E.; Kordás, K.; George, T.F.; Leppävuori, S. Thermal Oxidation of Porous Silicon: Study on Reaction Kinetics. J. Phys. Chem. B 2004, 108, 12744–12747. [Google Scholar] [CrossRef]
- Kumeria, T.; Wang, J.; Chan, N.; Harris, T.J.; Sailor, M.J. Visual Sensor for Sterilization of Polymer Fixtures Using Embedded Mesoporous Silicon Photonic Crystals. ACS Sens. 2018, 3, 143–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shtenberg, G.; Massad-Ivanir, N.; Fruk, L.; Segal, E. Nanostructured Porous Si Optical Biosensors: Effect of Thermal Oxidation on their Performance and Properties. ACS Appl. Mater. Interfaces 2014, 6, 16049–16055. [Google Scholar] [CrossRef]
- El-Gamal, A.A.; Ibrahim, S.M.; Amin, M. Impact of Thermal Oxidation on the Structural and Optical Properties of Porous Silicon Microcavity. Nanomater. Nanotechnol. 2017, 7, 184798041773570. [Google Scholar] [CrossRef] [Green Version]
- Martín-Sánchez, D.; Kovylina, M.; Ponce-Alcántara, S.; García-Rupérez, J. Thermo-Optic Coefficient of Porous Silicon in the Infrared Region and Oxidation Process at Low Temperatures. J. Electrochem. Soc. 2019, 166, B355–B359. [Google Scholar] [CrossRef]
- Wang, J.; Lee, G.Y.; Kennard, R.; Barillaro, G.; Bisiewicz, R.H.; Cortez Lemus, N.A.; Cao, X.C.; Anglin, E.J.; Park, J.S.; Potocny, A.; et al. Engineering the Properties of Polymer Photonic Crystals with Mesoporous Silicon Templates. Chem. Mater. 2017, 29, 1263–1272. [Google Scholar] [CrossRef]
- Vos, W.L.; Sprik, R.; van Blaaderen, A.; Imhof, A.; Lagendijk, A.; Wegdam, G.H. Strong Effects of Photonic Band Structures on the Diffraction of Colloidal Crystals. Phys. Rev. B Condens. Matter 1996, 53, 16231–16235. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Sun, H.-Y.; Yang, Y.-H.; Liu, L.-H.; Li, Z.-Y. Optical Properties and Color Generation Mechanism of Porous Anodic Alumina Films. Appl. Surf. Sci. 2011, 258, 1826–1830. [Google Scholar] [CrossRef]
- Olsen, T. On the Calculation of Power from Curvature of the Cornea. Br. J. Ophthalmol. 1986, 70, 152–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchinson, M.H.; Dorgan, J.R.; Knauss, D.M.; Hait, S.B. Optical Properties of Polylactides. J. Polym. Environ. 2006, 14, 119–124. [Google Scholar] [CrossRef]
- Bruggeman, D.A.G. Berechnung verschiedener physikalischer Konstanten von heterogenen Substanzen. I. Dielektrizitätskonstanten und Leitfähigkeiten der Mischkörper aus isotropen Substanzen. Ann. Phys. 1935, 416, 636–664. [Google Scholar] [CrossRef]
- Vichaibun, V.; Chulavatnatol, M. A New Assay for the Enzymatic Degradation of Polylactic Acid. Sci. Asia 2003, 29, 297. [Google Scholar] [CrossRef]
- Yamashita, K.; Kikkawa, Y.; Kurokawa, K.; Doi, Y. Enzymatic Degradation of Poly(L-Lactide) Film by Proteinase K: Quartz Crystal Microbalance and Atomic Force Microscopy Study. Biomacromolecules 2005, 6, 850–857. [Google Scholar] [CrossRef]
- Dordi, B.; Schönherr, H.; Vancso, G.J. Reactivity in the Confinement of Self-Assembled Monolayers: Chain Length Effects on the Hydrolysis of Self-Assembled Monolayers of N-Hydroxy-Succinimide Ester Disulfides of Gold. Langmuir 2003, 19, 5780–5786. [Google Scholar] [CrossRef]
- Pham, V.H.; Nguyen, T.V.; Nguyen, T.A.; Pham, V.D.; Bui, H. Nano Porous Silicon Microcavity Sensor for Determination Organic Solvents and Pesticide in Water. Adv. Nat. Sci. Nanosci. Nanotechnol. 2014, 5, 45003. [Google Scholar] [CrossRef]
- Salem, M.S.; Sailor, M.J.; Harraz, F.A.; Sakka, T.; Ogata, Y.H. Electrochemical Stabilization of Porous Silicon Multilayers for Sensing Various Chemical Compounds. J. Appl. Phys. 2006, 100, 83520. [Google Scholar] [CrossRef] [Green Version]
- Engel, L.S.; Hill, J.M.; Caballero, A.R.; Green, L.C.; O’Callaghan, R.J. Protease IV, a Unique Extracellular Protease and Virulence Factor from Pseudomonas aeruginosa. J. Biol. Chem. 1998, 273, 16792–16797. [Google Scholar] [CrossRef] [Green Version]
- Galdino, A.C.M.; Branquinha, M.H.; Santos, A.L.S.; Viganor, L. Pseudomonas aeruginosa and Its Arsenal of Proteases: Weapons to Battle the Host. In Pathophysiological Aspects of Proteases; Chakraborti, S., Dhalla, N., Eds.; Springer: Singapore, 2017; pp. 381–397. [Google Scholar] [CrossRef]
- Bowen, W.R.; Gan, Q. Properties of Microfiltration Membranes: The Effects of Adsorption and Shear on the Recovery of an Enzyme. Biotechnol. Bioeng. 1992, 40, 491–497. [Google Scholar] [CrossRef]
- Hernandez-Pinzon, I.; Millan, F.; Bautista, J. Study of the Loss of Streptokinase Activity during Cross-Flow Microfiltration: I. Immunologic Approach. Enzym. Microb. Technol. 1995, 17, 911–914. [Google Scholar] [CrossRef]
- Kaur, K.; Chelangat, W.; Druzhinin, S.I.; Karuri, N.W.; Müller, M.; Schönherr, H. Quantitative E. coli Enzyme Detection in Reporter Hydrogel-Coated Paper Using a Smartphone Camera. Biosensors 2021, 11, 25. [Google Scholar] [CrossRef]
- Haas, S.; Hain, N.; Raoufi, M.; Handschuh-Wang, S.; Wang, T.; Jiang, X.; Schönherr, H. Enzyme Degradable Polymersomes from Hyaluronic Acid-blockpoly(ε-caprolactone) Copolymers for the Detection of Enzymes of Pathogenic Bacteria. Biomacromolecules 2015, 16, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2019, 7, 7.2.16. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhusaini, Q.; Scheld, W.S.; Jia, Z.; Das, D.; Afzal, F.; Müller, M.; Schönherr, H. Bare Eye Detection of Bacterial Enzymes of Pseudomonas aeruginosa with Polymer Modified Nanoporous Silicon Rugate Filters. Biosensors 2022, 12, 1064. https://doi.org/10.3390/bios12121064
Alhusaini Q, Scheld WS, Jia Z, Das D, Afzal F, Müller M, Schönherr H. Bare Eye Detection of Bacterial Enzymes of Pseudomonas aeruginosa with Polymer Modified Nanoporous Silicon Rugate Filters. Biosensors. 2022; 12(12):1064. https://doi.org/10.3390/bios12121064
Chicago/Turabian StyleAlhusaini, Qasim, Walter Sebastian Scheld, Zhiyuan Jia, Dipankar Das, Faria Afzal, Mareike Müller, and Holger Schönherr. 2022. "Bare Eye Detection of Bacterial Enzymes of Pseudomonas aeruginosa with Polymer Modified Nanoporous Silicon Rugate Filters" Biosensors 12, no. 12: 1064. https://doi.org/10.3390/bios12121064