Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review
Abstract
:1. Introduction
- Biopolymers are macromolecules composed by the repetition of subunits coming from biological sources. Generally, biopolymers present lower chemical resistance processability and mechanical properties similar to those of synthetic polymers. These peculiarities, combined with their biodegradability efficiency, make biopolymers suitable to be employed in cosmetics [3], in food packaging [4] and in the production of novel medicine compounds [5]. Moreover, the use of biopolymers minimizes the employment of fossil fuels, which is crucial to prevent the undesirable release of greenhouse gases (GHG) during their manipulation and processing. The most promising biopolymers are based on plant cell wall constituents such as the plant cell walls, composed of lignin, cellulose and hemicelluloses, which are highly entangled.
- Hydrogels are three-dimensional network structures formed by flexible chains interconnected in set ways and swollen by liquid media. Hydrogels can undergo large and reversible expansion or shrinkage under specific conditions which confer their properties. Hydrogels are also employed for the realization of molecular sieves [6], glucose sensors [7], drug delivery systems [8], contact lenses [9], battery binders [10], disposable diapers [11] or bioinks for 3-D printing [12], among others.
- Dendrimers are three-dimensional branched polymeric macromolecules formed by arborescent construction. Conversely to polymers, where the molecular bond formation is probabilistic, the dendrimer molecule distribution is precise and the chemical bonds between atoms can be accurately described. Dendrimers treasure properties such as self-assembly, chemical stability, low cytotoxicity, polyvalency and good solubility. These properties are relevant in the developing of different fields such as molecular electronics [13], nanomedicine [14], light energy harvesting [15] and catalysis [16].
- Blends are constituted by homogeneous mixture of two or more polymers called homopolymers and copolymers, respectively, that have been mixed together to produce a new material with different physico-chemical properties. Blends have gained interest due to their ability to modify their mechanical properties. For this reason, blends are exploited for rubber toughening [17], food packaging [18], creation of supports for protein immobilization [19] or design of selective ion-exchange systems [20].
- Foams are formed by a gas–polymer mixture that provides microcellular structure with inner hollow pores. Foams can be rigid or flexible depending on the geometry of their inner structures. Thanks to their low density, high thermal and acoustic insulation and damping properties, foams are extensive applied for building construction [21], antipollution treatments [22], electronic shielding [23], fuel cells [24] and tissue engineering [25], among others.
- Liquid crystals are substances flowing like liquids and containing some degree of molecular arrangement ordering. Liquid crystals show excellent electro-optical, reflectance, anisotropic polarizability and low energy consumption qualities. Liquid crystals are widely used as detergent [26], for the realization of displays [27], thermal detection [28] and clinical diagnosis sensors [29].
- Finally, cells and bacteria are the fundamental anatomical unit of all living organisms and prokaryotic microorganisms that do not bear defined nuclei and generally internal membranous organelles, respectively. Cells can be presented as a biological source for regenerative medicine [30]. Bacteria are suitable as a prototype to fabricate microrobots due to their high motility and convenient controllability [31]. Moreover, the next generation of engineering bioreactors will be focused on cells and bacteria [32].
2. Mechanical Properties
3. Non-Nanotechnology Techniques to Determine Mechanical Properties
3.1. Multifrequency Magnetic Resonance Elastography (MRE)
3.2. Ultrasonic Pulse Testing
3.3. Tensile Testing
3.4. Indentation (Macroscale Level)
4. Mechanic Models to Ascertain Young’s Modulus
4.1. Hertz Model
4.2. Johnson, Kendall and Roberts (JKR) Model
4.3. Derjaguin, Müller and Toporov (DMT) Model
4.4. Maugis Model
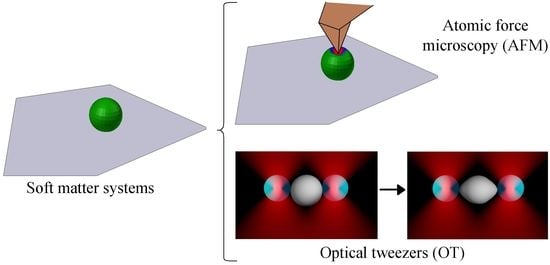
5. Working Principle of Nanotechnology Tools to Elicit Mechanical Properties
5.1. Atomic Force Microscopy (AFM)
5.2. Optical Tweezers (OT)
6. Recent Examples of Elastic Properties Addressed on Soft Matter Systems
6.1. Atomic Force Microscopy (AFM) to Evaluate Elastic Properties of Soft Matter
6.2. Elasticity of Soft Matter Systems Addressed by Optical Tweezers (OT)
7. Discussion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Guo, X.; Wu, J.; Fang, D.; Zhang, Y. Soft mechanical metamaterials with unusual swelling behavior and tunable stress-strain curves. Sci. Adv. 2018, 4, 8535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boey, F.Y.C.; Fuchs, H.; Chen, X. Nanotechnology with soft matter: From structures to functions. Small 2011, 7, 1275–1277. [Google Scholar] [CrossRef] [PubMed]
- Adamiak, K.; Kurzawa, M.; Sionkowska, A. Physicochemical performance of collagen modified by Melissa officinalis extract. Cosmetics 2021, 8, 95. [Google Scholar] [CrossRef]
- Wang, Y.; Borgatta, J.; White, J.C. Protecting foods with biopolymer fibres. Nat. Food 2022, 3, 402–403. [Google Scholar] [CrossRef]
- Baranwal, J.; Barse, B.; Fais, A.; Delogu, G.L.; Kumar, A. Biopolymer: A sustainable material for food and medical applications. Polymers 2022, 14, 983. [Google Scholar] [CrossRef]
- Ma, H.; Jiao, K.; Xu, X.; Song, J. Synthesis and characterization of a new aluminosilicate molecular sieve from aluminosilica perhydrate hydrogel. Materials 2020, 13, 5469. [Google Scholar] [CrossRef]
- Ando, M.; Tsuchiya, M.; Itai, S.; Murayama, T.; Kurashina, Y.; Heo, Y.J.; Onoe, H. Janus hydrogel microbeads for glucose sensing with pH calibration. Sensors 2021, 21, 4829. [Google Scholar] [CrossRef] [PubMed]
- Bordbar-Khiabini, A.; Gasik, M. Smart hydrogels for advanced drug delivery systems. Int. J. Mol. Sci. 2022, 23, 3665. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, M.; Wang, W.; Liu, H.; Deng, H.; Du, Y.; Shi, X. Electrodeposition induced covalent cross-linking of chitosan for electrofabrication of hydrogel contact lenses. Carbohydr. Polym. 2022, 292, 119678. [Google Scholar] [CrossRef]
- Xie, D.; Zhao, J.; Jiang, Q.; Wang, H.; Huang, H.; Rao, P.; Mao, J. A High-performance alginate hydrogel binder for aqueous Zn-Ion batteries. ChemPhysChem 2022, 23, e202200106. [Google Scholar] [CrossRef]
- Peng, N.; Wang, Y.; Ye, Q.; Liang, L.; An, Y.; Li, Q.; Chang, C. Biocompatible cellulose-based superabsorbent hydrogels with antimicrobial activity. Carbohydr. Polym. 2016, 137, 59–64. [Google Scholar] [CrossRef]
- Chimene, D.; Lennox, K.K.; Kaunas, R.K.; Gaharwar, A.K. Advanced bioinks for 3D printing: A materials science perspective. Ann. Biomed. Eng. 2016, 44, 2090–2102. [Google Scholar] [CrossRef] [PubMed]
- Lloveras, V.; Liko, F.; Muñoz-Gómez, J.L.; Veciana, J.; Vidal-Gancedo, J. Redox-active PTM radical dendrimers as promising multifunctional molecular switches. Chem. Mater. 2019, 31, 9400–9412. [Google Scholar] [CrossRef]
- Dias, A.P.; da Silva Santos, S.; da Silva, J.V.; Parise-Filho, R.; Ferreira, E.I.; El Seoud, O.; Giarolla, J. Dendrimers in the context of nanomedicine. Int. J. Pharm. 2020, 573, 118814. [Google Scholar] [CrossRef]
- Li, W.-J.; Wang, X.-Q.; Zhang, D.-Y.; Hu, Y.-X.; Xu, W.-T.; Xu, L.; Wang, W.; Yang, H.-B. Artificial light-harvesting systems based on AIEgen-branched rotaxane dendrimers for efficient photocatalysis. Angew. Chem. Int. Ed. Engl. 2021, 60, 18761–18768. [Google Scholar] [CrossRef]
- Karahanov, E.; Maximov, A.; Zolotukhina, A. Heterogeneous dendrimer-based catalysts. Polymers 2022, 14, 981. [Google Scholar] [CrossRef] [PubMed]
- Gigante, V.; Bosi, L.; Parlanti, P.; Gemmi, M.; Aliotta, L.; Lazzeri, A. Analysis of the damage mechanism around the crack tip for two rubber-toughened PLA-based blends. Polymers 2021, 13, 4053. [Google Scholar] [CrossRef]
- Arrieta, M.P.; Samper, M.D.; Aldas, M.; López, J. On the use of PLA-PHB blends for sustainable food packaging applications. Materials 2017, 10, 1008. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.S.; Yang, J.J.; Liu, C.X.; Bai, R.B. Immobilization of copper ions on chitosan/cellulose acetate blend hollow fiber membrane for protein adsorption. RSC Adv. 2017, 7, 10424–10431. [Google Scholar] [CrossRef] [Green Version]
- Son, T.Y.; Im, K.S.; Jung, H.N.; Nam, S.Y. Blended anion exchange membranes for vanadium redox flow batteries. Polymers 2021, 13, 2827. [Google Scholar] [CrossRef]
- Bedarf, P.; Dutto, A.; Zanini, M.; Dillenburger, B. Foam 3D printing for construction: A review of applications, materials, and processes. Autom. Constr. 2021, 130, 103861. [Google Scholar] [CrossRef]
- Vu, K.A.; Mulligan, C.N. Utilization of a biosurfactant foam/nanoparticle mixture for treatment of oil pollutants in soil. Environ. Sci. Pollut. Res. Int. 2022, 29, 88618–88629. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Zhang, M.; Liu, B.; Wang, F.; Wei, G.; Su, Z. Graphene foams for electromagnetic interference shielding: A Review. ACS Appl. Nano Mater. 2020, 3, 6140–6155. [Google Scholar] [CrossRef]
- Tan, W.C.; Saw, L.H.; Thiam, H.S.; Xuan, J.; Cai, Z.; Yew, M.C. Overview of porous media7metal foam application in fuel cells and solar power systems. Renew. Sustain. Energy Rev. 2018, 96, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Baino, F.; Tulyaganov, D.U.; Kahharov, Z.; Rahdar, A.; Verné, E. Foam-replicated diopside/fluorapatite/wollastonite-based blass-ceramic scaffolds. Ceramics 2022, 5, 120–130. [Google Scholar] [CrossRef]
- Oster, L.M.; Shechter, J.; Strain, B.; Shivrayan, M.; Thayumanavan, S.T.; Ross, J.L. Controlling liquid crystal configuration and phase using multiple molecular triggers. Molecules 2022, 27, 878. [Google Scholar] [CrossRef]
- A liquid crystal that could make your television screen brighter and clearer. Nature 2018, 557, 614. [CrossRef]
- Miskovic, V.; Malafronte, E.; Minetti, C.; Machrafi, H.; Varon, C.; Iorio, C.S. Thermotropic liquid crystals for temperature mapping. Front. Bioeng. Biotechnol. 2022, 10, 806362. [Google Scholar] [CrossRef]
- Ma, H.; Kang, Q.; Wang, T.; Xiao, J.; Yu, L. Liquid crystals-based sensor for the detection of lithocholic acid coupled with competitive host-guest inclusion. Colloids Surf. B Biointerfaces 2019, 173, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, X.; Zhang, Y.; Han, Y.; Chang, F.; Ding, J. Mesenchymal stem cells for regenerative medicine. Cells 2019, 8, 886. [Google Scholar] [CrossRef] [Green Version]
- Akolpoglu, M.B.; Alapan, Y.; Dogan, N.O.; Baltaci, S.F.; Yasa, O.; Tural, G.A.; Sitti, M. Magnetically steerable bacterial microrobots moving in 3D biological matrices for stimuli-responsive cargo delivery. Sci. Adv. 2022, 8, eabo6163. [Google Scholar] [CrossRef] [PubMed]
- Mouthuy, P.-A.; Snelling, S.; Hostettler, R.; Kharchenko, A.; Salmon, S.; Wainman, A.; Mimpen, J.; Paul, C.; Carr, A. Humanoid robots to mechanically stress human cells grown in soft bioreactors. Commun. Eng. 2022, 1, 2. [Google Scholar] [CrossRef]
- Menou, L.; Castelnovo, M. Mechanical stress relaxation in molecular self-assembly. Soft Matter 2019, 15, 6180–6189. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.T.; Wu, P.; Anariba, F. Modeling stress-strain nonlinear mechanics via entropy changes on surface wetting using the Born-Oppenheimer approximation. Results Eng. 2022, 13, 100349. [Google Scholar] [CrossRef]
- Erazo, O.; Vergara-Figueroa, J.; Valenzuela, P.; Gacitúa, W. Effect of the longitudinal tensile cree pon the stiffness of Radiata pine (Pinus radiata D. Don). Materials 2022, 15, 4314. [Google Scholar] [CrossRef]
- Ding, D.; Li, H.; Li, J.; Li, Z.; Yao, H.; Liu, L.; Tian, B.B.; Su, C.; Chen, F.; Shi, Y. Effect of mechanical forces on thermal stability reinforcement for lead based perovskite materials. J. Mater. Chem. A 2019, 7, 540–548. [Google Scholar] [CrossRef]
- Pustan, M.; Birleanu, C.; Voicu, R.; Muller, R. AFM characterization of temperature effect on the SU-8 mechanical and tribological properties. Polymers 2022, 14, 1009. [Google Scholar] [CrossRef]
- Jing, L.; Li, K.; Yang, H.; Chen, P.-Y. Recent advances in integration of 2D materials with soft matter for multifunctional robotic materials. Mater. Horiz. 2020, 7, 54–70. [Google Scholar] [CrossRef]
- Overvelde, J.T.B.; Weaver, J.C.; Hoberman, C.; Bertoldi, K. Rational design of reconfigurable prismatic architected materials. Nature 2017, 541, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Mazzoni, E.; Iaquinta, M.R.; Lanzillotti, C.; Mazziotta, C.; Maritati, M.; Montesi, M.; Sprio, S.; Tampieri, A.; Tognon, M.; Martini, F. Bioactive materials for soft tissue repair. Front. Bioeng. Biotechnol. 2021, 9, 613787. [Google Scholar] [CrossRef]
- Qu, R.; Li, G. Overview of liquid crystal biosensors: From basic theory to advanced applications. Biosensors 2022, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.Y.H.; Lee, C.S.; Pichika, M.R.; Cheng, S.F.; Lam, K.Y. PH responsive polyurethane for the advancement of biomedical and drug delivery. Polymers 2022, 14, 1672. [Google Scholar] [CrossRef]
- Berzin, F.; Lemkhanter, L.; Marcuello, C.; Chabbert, B.; Aguié-Béghin, V.; Molinari, M.; Castellani, R.; Vergnes, B. Influence of the polarity of the matrix on the breakage mechanisms of lignocellulosic fibers during twin-screw extrusión. Polym. Compos. 2020, 41, 1106–1117. [Google Scholar] [CrossRef]
- Li, S.; Dong, S.; Xu, W.; Tu, S.; Yan, L.; Zhao, C.; Ding, J.; Chen, X. Antibacterial hydrogels. Adv. Sci. 2018, 5, 1700527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, V.; Ifraimov, N.; Wylie, R.G. Modulating the thermoresponse of polymer-protein conjugates with hydrogels for controlled release. Polymers 2021, 13, 2772. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, W.; Zhao, X.; Zhao, Y. Synthesis and thermoresponsive behaviors of thermo-, pH-, CO2-, and oxidation-responsive linear and cyclic graft copolymers. Macromolecules 2017, 50, 3411–3423. [Google Scholar] [CrossRef]
- Marturano, V.; Bizarro, V.; Ambrogi, V.; Cutignano, A.; Tommonaro, G.; Abbamondi, G.R.; Giamberini, M.; Tylkowski, B.; Carfagna, C.; Cerruti, P. Light-responsive nanocapsule-coated polymer films for antimicrobial active packaging. Polymers 2019, 11, 68. [Google Scholar] [CrossRef] [Green Version]
- Xiang, T.; Lu, T.; Zhao, W.-F.; Zhao, C.-S. Ionic-strength responsive zwitterionic copolymer hydrogels with tunable swelling and adsorption behaviors. Langmuir 2019, 35, 1146–1155. [Google Scholar] [CrossRef]
- Yang, D.-Q.; Chen, J.-H.; Cao, Q.-T.; Duan, B.; Chen, H.-J.; Yu, X.-C.; Xiao, Y.-F. Operando monitoring transition dynamics of responsive polymer using optofluidic microcavities. Light Sci. Appl. 2021, 10, 128. [Google Scholar] [CrossRef]
- Spalek, J.; Ociepa, P.; Deptula, P.; Piktel, E.; Daniluk, T.; Król, G.; Góźdź, S.; Bucki, R.; Okla, S. Biocompatible materials in otorhinolaryngology and their antibacterial properties. Int. J. Mol. Sci. 2022, 23, 2575. [Google Scholar] [CrossRef]
- Streitberger, K.-J.; Reiss-Zimmermann, M.; Freimann, F.B.; Bayerl, S.; Guo, J.; Arlt, F.; Wuerfel, J.; Braun, J.; Hoffmann, K.-T.; Sack, I. High-resolution mechanical imaging of glioblastoma by multifrequency magnetic resonance elastography. PLoS ONE 2014, 9, e110588. [Google Scholar] [CrossRef]
- Khoury, M.; Tourtollet, G.E.; Schröder, A. Contactless measurement of the elastic Young’s modulus of paper by an ultrasonic technique. Ultrasonics 1999, 37, 133–139. [Google Scholar] [CrossRef]
- Lopez, D.M.B.; Ahmad, R. Tensile mechanical behaviour of multi-polymer sandwich structures via fused deposition modelling. Polymers 2020, 12, 651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalcioglu, Z.I.; Mahmoodian, R.; Hu, Y.; Suo, Z.; Van Vliet, K.J. From macro-to microscale poroelastic characterization of polymeric hydrogels via indentation. Soft Matter 2012, 8, 3393–3398. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, D.J.; Dumitru, A.C.; Lo Giudice, C.; Gaub, H.E.; Hinterdorfer, P.; Hummer, G.; De Yoreo, J.J.; Dufrêne, Y.F.; Alsteens, D. Atomic force microscopy-based force spectroscopy and multiparametric imaging of biomolecular and cellular systems. Chem. Rev. 2021, 121, 11701–11725. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.; Ferreira, P.; Marcuello, C.; Usón, A.; Miramar, M.D.; Peleato, M.L.; Lostao, A.; Susin, S.A.; Medina, M. Key residues regulating the reductase activity of the human mitochondrial apoptosis inducing factor. Biochemistry 2015, 54, 5175–5184. [Google Scholar] [CrossRef] [PubMed]
- Sebastián, M.; Lira-Navarrete, E.; Serrano, A.; Marcuello, C.; Velázquez-Campoy, A.; Lostao, A.; Hurtado-Guerrero, R.; Medina, M.; Martínez-Júlvez, M. The FAD synthetase from the human pathogen Streptococcus pneumoniae: A bifunctional enzyme exhibiting activity-dependent redox requirements. Sci. Rep. 2017, 7, 7609. [Google Scholar] [CrossRef] [Green Version]
- Marcuello, C.; Frempong, G.A.; Balsera, M.; Medina, M.; Lostao, A. Atomic force microscopy to elicit conformational transitions of ferredoxin-dependent flavin thioredoxin reductases. Antioxidants 2021, 10, 1437. [Google Scholar] [CrossRef]
- Marcuello, C.; de Miguel, R.; Martínez-Júlvez, M.; Gómez-Moreno, C.; Lostao, A. Mechanostability of the single-electron-transfer complexes of Anabaena Ferredoxin-NADP(+) reductase. ChemPhysChem 2015, 16, 3161–3169. [Google Scholar] [CrossRef]
- Pérez-Domínguez, S.; Caballero-Mancebo, S.; Marcuello, C.; Martínez-Júlvez, M.; Medina, M.; Lostao, A. Nanomechanical study of enzyme: Coenzyme complexes: Bipartite sites in plastidic Ferredoxin-NADP+ reductase for the interaction with NADP. Antioxidants 2022, 11, 537. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Rojo, R.; Marcuello, C.; Lostao, A.; Gómez-Moreno, C.; Mazo, J.J.; Falo, F. A physical picture for mechanical dissociation of biological complexes: From forces to free energies. Phys. Chem. Chem. Phys. 2017, 19, 4567–4575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcuello, C.; de Miguel, R.; Lostao, A. Molecular recognition of proteins through quantitative force maps at single molecule level. Biomolecules 2022, 12, 594. [Google Scholar] [CrossRef] [PubMed]
- Wieland, K.; Ramer, G.; Weiss, V.U.; Allmaier, G.; Lendl, B.; Centrone, A. Nanoscale chemical imaging of individual, chemotherapeutic cytarabine-loaded liposomal nanocarriers. Nano Res. 2019, 12, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, C.; Chambel, L.; Rodrigues, M.S.; Ferreira, L.P.; Cruz, M.M. Magnetotactic bacteria: Magnetism beyond magnetosomes. IEEE Trans. Nanobiosci. 2018, 17, 555–559. [Google Scholar] [CrossRef]
- Mair, L.O.; Chowdhury, S.; Paredex-Juarez, G.A.; Guix, M.; Bi, C.; Johnson, B.; English, B.W.; Jafari, S.; Baker-McKee, J.; Watson-Daniels, J.; et al. Magnetically aligned nanorods in alginate capsules (MANiACs): Soft matter tumbling robots for manipulation in drug delivery. Micromachines 2019, 10, 230. [Google Scholar] [CrossRef] [Green Version]
- Barra, A.; Alves, Z.; Ferreira, N.M.; Martins, M.A.; Oliveira, H.; Ferreira, L.P.; Cruz, M.M.; de Deus Carvalho, M.; Neumayer, S.M.; Rodriguez, B.J.; et al. Biocompatible chitosan-based composites with properties suitable for hyperthermia therapy. J. Mater. Chem. B 2020, 8, 1256–1265. [Google Scholar] [CrossRef]
- Shi, X.; Qing, W.; Marhaba, T.; Zhang, W. Atomic force microscopy—Scanning electrochemical microscopy (AFM-SECM) for nanoscale topographical and electrochemical characterization: Principles, applications and perspectives. Electrochim. Acta 2020, 332, 135472. [Google Scholar] [CrossRef]
- Pumera, M.; Sánchez, S.; Ichinose, I.; Tang, J. Electrochemical nanobiosensors. Sens. Actuators B Chem. 2007, 123, 1195–1205. [Google Scholar] [CrossRef]
- Ceballos-Laita, L.; Marcuello, C.; Lostao, A.; Calvo-Begueria, L.; Velázquez-Campoy, A.; Bes, M.T.; Fillat, M.F.; Peleato, M.-L. Microcystin-LR binds iron, and iron promotes self-assembly. Environ. Sci. Technol. 2017, 51, 4841–4850. [Google Scholar] [CrossRef]
- Bammes, B.E.; Jakana, J.; Schmid, M.F.; Chiu, W. Radiation damage effects at four specimen temperatures from 4 to 100 K. J. Struct. Biol. 2010, 169, 331–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, J.R.; Horne, R.W. Negative staining: A brief assessment of current technical benefits, limitations and future possibilities. Micron 1994, 25, 5–13. [Google Scholar] [CrossRef]
- Pool, R. Trapping with optical tweezers. Science 1988, 241, 1042. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Alshareedah, I.; Wang, W.; Ngo, J.; Moosa, M.M.; Banerjee, P.R. Molecular crowding tunes materials states of ribonucleoprotein condensates. Biomolecules 2019, 9, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustamante, C.; Alexander, L.; Maciuba, K.; Kaiser, C.M. Single-molecule studies of protein folding with optical tweezers. Annu. Rev. Biochem. 2020, 89, 443–470. [Google Scholar] [CrossRef]
- Blázquez-Castro, A.; Fernández-Piqueras, J.; Santos, J. Genetic material manipulation and modification by optical trapping and nanosurgery—A perspective. Front. Bioeng. Biotechnol. 2020, 8, 580937. [Google Scholar] [CrossRef]
- Shergill, B.; Meloty-Kapella, L.; Musse, A.A.; Weinmaster, G.; Botvinick, E. Optical tweezers studies on Notch: Single-molecule interaction strength is independent of ligand endocytosis. Dev. Cell 2012, 22, 1313–1320. [Google Scholar] [CrossRef] [Green Version]
- Meng, K.; Yang, H.; Wang, Y.; Sun, D. Modeling and control of single-cell migration induced by a chemoattractant-loaded microbead. IEEE Trans. Cybern. 2019, 49, 427–439. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Zou, R.; Hu, H.; Wang, Y.; Wang, F.; Ju, L.A. Recent advances of optical tweezers-based dynamic force spectroscopy and mechanical measurement assays for live-cell mechanobiology. Front. Phys. 2022, 10, 771111. [Google Scholar] [CrossRef]
- Vasse, G.F.; Buzón, P.; Melgert, B.N.; Roos, W.H.; van Rijn, P. Single cell reactomics: Real-time single-cell activation kinetics of optically trapped macrophages. Small Methods 2021, 5, e2000849. [Google Scholar] [CrossRef]
- Nieminen, T.A.; Loke, V.L.Y.; Stilgoe, A.B.; Knöner, G.; Brańczyk, A.M.; Heckenberg, N.R.; Rubinsztein-Dunlop, H. Optical tweezers computational toolbox. J. Opt. A Pure Appl. Opt. 2007, 9, S196. [Google Scholar] [CrossRef] [Green Version]
- Pesce, G.; Volpe, G.; Volpe, G.; Saso, A. Long-term influence of fluid inertia on the diffusion of a Brownian particle. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2014, 90, 042309. [Google Scholar] [CrossRef] [Green Version]
- Hooke, R. Lectures De Potentia Restitutiva, or of Spring. Explaining the Power of Springing Bodies; Martyn, J., Ed.; Royal Society: London, UK, 1678. [Google Scholar]
- Mohs, F.; Haidinger, W. Treatise on Mineralogy, or, The Natural History of the Mineral Kingdom. Landmarks of Science; A. Constable: London, UK, 1825. [Google Scholar] [CrossRef]
- Smith, R.L.; Sandland, G.E. An accurate method of determining the hardness of metals, with particular reference to those of a high degree of hardness. Proc. Inst. Mech. Eng. 1922, 1, 623–643. [Google Scholar] [CrossRef]
- Knoop, F.; Peters, C.G.; Emerson, W.B. A sensitive pyramidal-diamond tool for indentation measurements. J. Res. Natl. Bur. Stand. 1939, 23, 39–61. [Google Scholar] [CrossRef]
- Nohel, J.A. A nonlinear volterra equation in viscoelasticity. In Nonlinear Phenomena in Mathematical Sciences, Proceedings of the International Conference on Nonlinear Phenomena in Mathematical Sciences, Arlington, TX, USA, 16–20 June 1980; Academic Press: Cambridge, MA, USA, 1982; p. 747. [Google Scholar] [CrossRef]
- Schapery, R. Nonlinear viscoelastic and viscoplastic constitutive equations with growing damage. Int. J. Fract. 1999, 97, 33–66. [Google Scholar] [CrossRef]
- Chen, Y.; Smith, L.V. A nonlinear viscoelastic-viscoplastic model for adhesives. Mech. Time-Depend. Mater. 2021, 25, 565–579. [Google Scholar] [CrossRef]
- Nair, T.M.; Kumaran, M.G.; Unnikrishnan, G.; Pillai, V.B. Dynamic mechanical analysis of ethylene-propylene-diene monomer rubber and styrene-butadiene rubber blends. J. Appl. Polym. Sci. 2008, 112, 72–81. [Google Scholar] [CrossRef]
- Irwing, G.R. Linear fracture mechanics, fracture transition, and fracture control. Eng. Fract. Mech. 1968, 1, 241–257. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Feng, Y.; Wang, F.; Jeremić, B. Energy dissipation in solids due to material inelasticity, viscous coupling, and algorithmic damping. J. Energy. Mech. 2019, 145, 04019060. [Google Scholar] [CrossRef]
- McIlvain, G.; Ganji, E.; Cooper, C.; Killian, M.L.; Ogunnaike, B.A.; Johnson, C.L. Reliable preparation of agarose phantoms for use in quantitative magnetic resonance elastography. J. Mech. Behav. Biomed. Mater. 2019, 97, 65–73. [Google Scholar] [CrossRef]
- Chatelin, S.; Charpentier, I.; Corbin, N.; Meylheuc, L.; Vappou, J. An automatic differentiation-based gradient method for inversion of the shear wave equation in magnetic resonance elastography: Specific application in fibrous soft tissues. Phys. Med. Biol. 2016, 61, 5000–5019. [Google Scholar] [CrossRef] [PubMed]
- Snellings, J.; Keshi, E.; Tang, P.; Daneshgar, A.; Willma, E.C.; Haderer, L.; Klein, O.; Krenzien, F.; Malinka, T.; Asbach, P.; et al. Solid fraction determines stiffness and viscosity in decellularized pancreatic tissues. Biomater. Adv. 2022, 139, 212999. [Google Scholar] [CrossRef] [PubMed]
- Herthum, H.; Shahryari, M.; Tzschãtzsch, H.; Schrank, F.; Warmuth, C.; Gõrner, S.; Hetzer, S.; Neubauer, H.; Pfeuffer, J.; Braun, J.; et al. Real-Time multifrequency MR elastography of the human brain reveals rapid changes in viscoelasticity in response to the valsalva maneuver. Front. Bioeng. Biotechnol. 2021, 9, 666456. [Google Scholar] [CrossRef]
- Reiter, R.; Loch, F.N.; Kamphues, C.; Bayerl, C.; Marticorena García, S.R.; Siegmund, B.; Kühl, A.A.; Hamm, B.; Braun, J.; Sack, I.; et al. Feasibility of intestinal MR elastography in inflammatory bowel disease. J. Magn. Reson. Imaging 2022, 55, 815–822. [Google Scholar] [CrossRef]
- Grasland-Mongrain, P.; Zorgani, A.; Nakagawa, S.; Bernard, S.; Paim, L.G.; Fitzharris, G.; Catheline, S.; Cloutier, G. Ultrafast imaging of cell elasticity with optical microelastography. Proc. Natl. Acad. Sci. USA 2018, 115, 861–866. [Google Scholar] [CrossRef] [Green Version]
- Kino, G.S. Acoustic waves. In Devices, Imaging, and Analog Signal Processing; Prentice Hall: Upper Saddle River, NJ, USA, 1986. [Google Scholar]
- Judawisastra, H.; Claudia; Sasmita, F.; Agung, T.P. Elastic modulus determination of thermoplastic polymers with pulse-echo method ultrasonic testing. IOP Conf. Ser. Mater. Sci. Eng. 2019, 547, 012047. [Google Scholar] [CrossRef] [Green Version]
- Neumann, A.J.; Quinn, T.; Bryant, S.J. Nondestructive evaluation of a new hydrolytically degradable and photo-clickable PEG hydrogel for cartilage tissue engineering. Acta Biomater. 2016, 39, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Oh, S.; Wang, X.-Y.; Lin, R.-S. Hydration-strength-workability-durability of binary, ternary, and quaternary composite pastes. Materials 2021, 15, 204. [Google Scholar] [CrossRef]
- Dawood, E.T.; Mahammad, Y.Z.; Abbas, W.A.; Mannan, M.A. Toughness, elasticity and physical properties for the evaluation of foamed concrete reinforced with hybrid fibers. Heliyon 2018, 14, e01103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-C.; Wang, H.-Y.; Chen, C.-H.; Huang, C. Prediction of compressive strength using ultrasonic pulse velocity for CLSM with waste LCD glass concrete. J. Civ. Eng. Archit. 2015, 9, 691–700. [Google Scholar] [CrossRef] [Green Version]
- Abu-Nab, A.K.; Mohamed, K.G.; Abu-Bakr, A. Microcavitation dynamics in viscoelastic tissue during histotripsy process. J. Phys. Condens. Matter 2022, 34, 304005. [Google Scholar] [CrossRef]
- Baron, C.; Nguyen, V.-H.; Naili, S.; Guivier-Curien, C. Interaction of ultrasound waves with bone remodelling: A multiscale computational study. Biomech. Model Mechanobiol. 2020, 19, 1755–1764. [Google Scholar] [CrossRef]
- Miller, D.L.; Dong, Z.; Dou, C.; Patterson, B.; Raghavendran, K. Pulmonary capillary hemorrhage induced by acoustic radiation force impulse shear wave elastography in ventilated rats. J. Ultrasound Med. 2019, 38, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Hollomon, J.H. Tensile deformation. Trans. Metall. Soc. AIME 1945, 162, 268–290. [Google Scholar]
- Bowen, A.W.; Partridge, P.G. Limitations of the Hollomon strain-hardening equation. J. Phys. D Appl. Phys. 1974, 7, 969. [Google Scholar] [CrossRef]
- Swift, H.W. Plastic instability under plane stress. J. Mech. Phys. Solids 1952, 1, 1–18. [Google Scholar] [CrossRef]
- Voce, E. The relationship between stress and strain for homogeneous deformations. J. Inst. Metals 1948, 74, 537–562. [Google Scholar]
- Harussani, M.M.; Sapuan, S.M.; Firdaus, A.H.M.; El-Badry, Y.A.; Hussein, E.E.; El-Bahy, Z.M. Determination of the tensile properties and biodegradability of cornstarch-based biopolymers plasticized with sorbitol and glycerol. Polymers 2021, 13, 3709. [Google Scholar] [CrossRef]
- Osman, A.F.; Siah, L.; Alrashdi, A.A.; Ul-Hamid, A.; Ibrahim, I. Improving the tensile and tear properties of thermoplastic starch/dolomite biocomposite film through sonication process. Polymers 2021, 13, 274. [Google Scholar] [CrossRef]
- Gerbin, E.; Frapart, Y.-M.; Marcuello, C.; Cottyn, B.; Foulon, L.; Pernes, M.; Crônier, D.; Molinari, M.; Chabbert, B.; Ducrot, P.-H.; et al. Dual antioxidant properties and organic radical stabilization in cellulose nanocomposite films functionalized by in situ polymerization of coniferyl alcohol. Biomacromolecules 2020, 21, 3163–3175. [Google Scholar] [CrossRef] [PubMed]
- Gerbin, E.; Rivière, G.N.; Foulon, L.; Frapart, Y.M.; Cottyn, B.; Pernes, M.; Marcuello, C.; Godon, B.; Gainvors-Claisse, A.; Crônier, D.; et al. Tuning the functional properties of Lignocellulosic films by controlling the molecular and supramolecular structure of lignin. Int. J. Biol. Macromol. 2021, 181, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, Y.; Wang, J.; Zhu, Y.; Zhu, Y.; Zhang, X.; Liao, J.; Li, X.; Wu, X.; Qin, Y.-X.; et al. A composite hydrogel with high mechanical strength, fluorescence, and degradable behavior for bone tissue engineering. Polymers 2019, 11, 1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, Q.; Liu, H.; Yang, Y. A highly mechanical, conductive, and cryophylactic double network hydrogel for flexible and low-temperature tolerant strain sensors. Gels 2022, 8, 424. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, J. Natural rubber/dendrimer modified montmorillonite nanocomposites: Mechanical and flame-retardant properties. Materials 2017, 11, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Wang, X. Chlorhexidine-loaded poly /amido amine) dendrimer and a dental adhesive containing amorphous calcium phosphate nanofillers for enhancing bonding durability. Dent. Mater. 2022, 38, 824–834. [Google Scholar] [CrossRef]
- Embabi, M.; Kweon, M.S.; Chen, Z.; Lee, P.C. Tunable tensile properties of polypropylene and polyethylene terephthalate fibrillar blends through micro-/nanolayered extrusion technology. Polymers 2020, 12, 2585. [Google Scholar] [CrossRef]
- Pangesty, A.I.; Todo, M. Improvement of mechanical strength of tissue engineering scaffold due to the temperature control of polymer blend solution. J. Funct. Biomater. 2021, 12, 47. [Google Scholar] [CrossRef]
- Kordován, M.Á.; Hegedüs, C.; Czifrák, K.; Lakatos, C.; Kálmán-Szabó, I.; Daróczi, L.; Zsuga, M.; Kéki, S. Novel Polyurethane scaffolds containing sucrose crosslinker for dental application. Int. J. Mol. Sci. 2022, 23, 7904. [Google Scholar] [CrossRef]
- Ter-Zakaryan, K.A.; Zhukov, A.D.; Bobrova, E.Y.; Bessonov, I.V.; Mednikova, E.A. Foam polymers in multifunctional insulating coatings. Polymers 2021, 13, 3698. [Google Scholar] [CrossRef]
- Liu, Z.; Xiong, Y.; Hao, J.; Zhang, H.; Cheng, X.; Wang, H.; Chen, W.; Zhou, C. Liquid crystal-based organosilicone elastomers with supreme mechanical adaptability. Polymers 2022, 14, 789. [Google Scholar] [CrossRef]
- Pillet, B.; Badel, P.; Pierrat, B. Effects of cryo-preservation of skeletal muscle tissues mechanical behavior under tensile and peeling tests until ruptura. J. Mech. Behav. Biomed. Mater. 2022, 132, 105273. [Google Scholar] [CrossRef] [PubMed]
- Pangesty, A.I.; Arahira, T.; Todo, M. Characterization of tensile mechanical behavior of MSCs/PLCL hybrid layered sheet. J. Funct. Biomater. 2016, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Zhao, D.; Tian, Y.; Wang, S.; Zhao, H.; Zhang, J. Study on the deformation mechanism of spherical diamond indenter and its influence on 3C-SiC sample during nanoindentation process via molecular dynamics simulation. Mater. Sci. Semicond. Process. 2019, 90, 143–150. [Google Scholar] [CrossRef]
- Schneider, J.-M.; Bigerelle, M.; Iost, A. Statistical analysis of the Vickers hardness. Mater. Sci. Eng. A 1999, 262, 256–263. [Google Scholar] [CrossRef]
- Oliver, W.C.; Pharr, G.M. Measurement of hardness and elastic modulus by instrumented indentation: Advances in understanding and refinements to methodology. J. Mater. Res. 2004, 19, 3. [Google Scholar] [CrossRef]
- Iqbal, T.; Camargo Jr, S.S.; Yasin, S.; Farooq, U.; Shakeel, A. Nano-indentation response of ultrahigh molecular weight polyethylene (UHMWPE): A detailed analysis. Polymers 2020, 12, 795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, L.; Fonseca, R.; Pinto, L.F.V.; Ferreira, F.C.; Almeida, A.; Rodrigues, A. Strategy to improve the mechanical properties of bioabsorbable materials based on chitosan for orthopedic fixation applications. J. Mech. Behav. Biomed. Mater. 2020, 103, 103572. [Google Scholar] [CrossRef]
- Major, R.; Trembecka-Wójciga, K.; Kot, M.; Lackner, J.M.; Wilczek, P.; Major, B. In vitro hemocompatibility on thin ceramic and hydrogel films deposited on polymer substrate performed in arterial flow conditions. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 61, 15–22. [Google Scholar] [CrossRef]
- Bano, S.; Iqbal, T.; Ramzan, N.; Farooq, U. Study of surface mechanical characteristics of ABS/PC blends using nanoindentation. Processes 2021, 9, 637. [Google Scholar] [CrossRef]
- Zhang, Z.; Bellisario, D.; Quadrini, F.; Jestin, S.; Ravanelli, F.; Castello, M.; Li, X.; Dong, H. Nanoindentation of multifunctional smart composites. Polymers 2022, 14, 2945. [Google Scholar] [CrossRef]
- Riad, K.B.; Hoa, S.V.; Wood-Adams, P.M. Photocuring graphene oxide liquid crystals for high-strength structural materials. ACS Omega 2022, 7, 21192–21198. [Google Scholar] [CrossRef] [PubMed]
- Schwiedrzik, J.J.; Zysset, P.K. The influence of yield surface shape and damage in the depth-dependent response of bone tissue to nanoindentation using spherical and Berkovich indenters. Comput. Methods Biomech. Biomed. Engin. 2013, 18, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Hertz, H. Ueber die Berührung fester elastischer Körper. J. Reine Angew. Math. 1882, 92, 156–171. [Google Scholar] [CrossRef]
- Rigato, A.; Miyagi, A.; Scheuring, S.; Rico, F. High-frequency microrheology reveals cytoskeleton dynamics in living cells. Nat. Phys. 2017, 13, 771–775. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.L.; Kendall, K.; Roberts, A.D. Surface energy and the contact of elastic solids. Proc. R. Soc. Lond. A Math. Phys. Sci. 1971, 324, 301–313. [Google Scholar] [CrossRef] [Green Version]
- Derjaguin, B.V.; Muller, V.M.; Toporov, Y.P. Effect of contact deformations on the adhesion of particles. J. Colloid Interf. Sci. 1975, 53, 314–326. [Google Scholar] [CrossRef]
- Maugis, D. Adhesion of spheres: The JKR-DMT transition using a dugdale model. J. Colloid Interf. Sci. 1992, 150, 243–269. [Google Scholar] [CrossRef]
- Bradley, R.S. LXXIX. The cohesive force between solid surfaces and the surface energy of solids. Lond. Edin. Dubl. Phil. Mag. J. Sci. 1932, 13, 853–862. [Google Scholar] [CrossRef]
- Schillers, H.; Rianna, C.; Schäppe, J.; Luque, T.; Doschke, H.; Wälter, M.; Uriarte, J.J.; Campillo, N.; Michanetzis, G.P.A.; Bobrowska, J.; et al. Standardized nanomechanical atomic force microscopy procedure (SNAP) for measuring soft and biological samples. Sci. Rep. 2017, 7, 5117. [Google Scholar] [CrossRef] [Green Version]
- Ambaum, M.H.P. Chapter 2—The first and second laws. In Thermal Physics of the Atmosphere, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 17–38. [Google Scholar] [CrossRef]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef] [Green Version]
- Sader, J.E. Method for the calibration of atomic force microscope cantilevers. Rev. Sci. Instrum. 1995, 66, 3789. [Google Scholar] [CrossRef] [Green Version]
- Ashkin, A. Acceleration and trapping of particles by radiation pressure. Phys. Rev. Lett. 1970, 24, 156–159. [Google Scholar] [CrossRef] [Green Version]
- Ashkin, A.; Dziedzic, J.M.; Bjorkholm, J.E.; Chu, S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 1986, 11, 288–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashkin, A.; Dziedzic, J.M.; Yamane, T. Optical trapping and manipulation of single cells using infrared laser beams. Nature 1987, 330, 769–771. [Google Scholar] [CrossRef]
- Ashkin, A. Atomic-beam deflection by resonance-radiation pressure. Phys. Rev. Lett. 1970, 25, 1321–1324. [Google Scholar] [CrossRef]
- Maragò, O.M.; Jones, P.H.; Gucciardi, P.G.; Volpe, G.; Ferrari, A.C. Optical trapping and manipulation of nanostructures. Nat. Nanotechnol. 2013, 8, 807–819. [Google Scholar] [CrossRef] [Green Version]
- Dholakia, K.; Čižmár, T. Shaping the future of manipulation. Nat. Photonics 2011, 5, 335–342. [Google Scholar] [CrossRef]
- Ashkin, A.; Dziedzic, J.M. Optical trapping and manipulation of viruses and bacteria. Science 1987, 235, 1517–1520. [Google Scholar] [CrossRef]
- Fazal, F.M.; Block, S.M. Optical tweezers study life under tension. Nat. Photonics 2011, 5, 318–321. [Google Scholar] [CrossRef]
- Volpe, G.; Marago, O.M.; Rubinsztein-Dunlop, H.; Pesce, G.; Stilgoe, A.; Volpe, G.; Tkachenko, G.; Truong, V.G.; Chormaic, S.N.; Kalantarifard, F. Roadmap for Optical Tweezers. J. Phys. Photonics 2023. [Google Scholar] [CrossRef]
- Jones, P.H.; Maragò, O.M.; Volpe, G. Optical Tweezers: Principles and Applications; Cambridge University Press: Cambridge, UK, 2015. [Google Scholar] [CrossRef]
- Mischenko, M.I.; Travis, L.D.; Lacis, A.A. Scattering, Absorption, and Emission of Light by Small Particles; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Born, M.; Wolf, E.; Bhatia, A.B.; Clemmow, P.C.; Gabor, D.; Stokes, A.R.; Taylor, A.M.; Wayman, P.A.; Wilcock, W.L. Principles of Optics: Electromagnetic Theory of Propagation, Interference and Diffraction of Light, 7th ed.; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar] [CrossRef]
- Magazzù, A.; Spadaro, D.; Donato, M.G.; Sayed, R.; Messina, E.; D’Andrea, C.; Foti, A.; Fazio, B.; Iatí, M.A.; Irrera, A.; et al. Optical tweezers: A non-destructive tool for soft and biomaterial investigations. Rend. Fis. Acc. Lincei. 2015, 26, 203–218. [Google Scholar] [CrossRef]
- Polimeno, P.; Magazzù, A.; Iatí, M.A.; Patti, F.; Saija, R.; Boschi, C.D.E.; Donato, M.G.; Gucciardi, P.G.; Jones, P.H.; Volpe, G.; et al. Optical tweezers and their applications. J. Quant. Spectrosc. Radiat. Transf. 2018, 218, 131–150. [Google Scholar] [CrossRef] [Green Version]
- Gieseler, J.; Deutsch, B.; Quidant, R.; Novotny, L. Subkelvin parametric feedback cooling of a laser-trapped nanoparticle. Phys. Rev. Lett. 2012, 109, 103603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, R.; Vollmer, W.; Perry, J.D.; Stoodley, P.; Chen, J. Simultaneous determination of the mechanical properties and turgor of a single bacterial cell using atomic force microscopy. Nanoscale 2022, 14, 12060–12068. [Google Scholar] [CrossRef]
- Francius, G.; Polyakov, P.; Merlin, J.; Abe, Y.; Ghigo, J.-M.; Merlin, C.; Beloin, C.; Duval, J.F.L. Bacterial surface appendages strongly impact nanomechanical and electrokinetic properties of Escherichia coli cells subjected to osmotic stress. PLoS ONE 2011, 6, e20066. [Google Scholar] [CrossRef]
- Calò, A.; Romin, Y.; Srouji, R.; Zambirinis, C.P.; Fan, N.; Santella, A.; Feng, E.; Fujisawa, S.; Turkekul, M.; Huang, S. Spatial mapping of the collagen distribution in human and mouse tissues by force volume atomic force microscopy. Sci. Rep. 2020, 10, 15664. [Google Scholar] [CrossRef]
- Roos, W.H.; Gertsman, I.; May, E.R.; Brooks, C.L., 3rd; Johnson, J.E.; Wuite, G.J.L. Mechanics of bacteriophage maturation. Proc. Natl. Acad. Sci. USA 2012, 109, 2342–2347. [Google Scholar] [CrossRef] [Green Version]
- Jeon, B.; Jung, H.G.; Lee, S.W.; Lee, G.; Shim, J.H.; Kim, M.O.; Kim, B.J.; Kim, S.-H.; Lee, H.; Lee, S.W.; et al. Melanoma detection by AFM indentation of histological specimens. Diagnostics 2022, 12, 1736. [Google Scholar] [CrossRef]
- Chen, X.; Hughes, R.; Mullin, N.; Hawkins, R.J.; Holen, I.; Brown, N.J.; Hobbs, J.K. Mechanical heterogeneity in the bone microenvironment as characterized by atomic force microscopy. Biophys. J. 2020, 119, 502–513. [Google Scholar] [CrossRef]
- Marcuello, C.; Foulon, L.; Chabbert, B.; Aguié-Béghin, V.; Molinari, M. Atomic force microscopy reveals how relative humidity impacts the Young’s modulus of lignocellulosic polymers and their adhesion with cellulose nanocrystals at the nanoscale. Int. J. Biol. Macromol. 2020, 147, 1064–1075. [Google Scholar] [CrossRef]
- Bouali, A.B.; Montembault, A.; David, L.; Von Boxberg, Y.; Viallon, M.; Hamdi, B.; Nothias, F.; Fodil, R.; Féréol, S. Nanoscale mechanical properties of chitosan hydrogels as revealed by AFM. Prog. Biomater. 2020, 9, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Miras, J.; Liu, C.; Blomberg, E.; Thormann, E.; Vílchez, S.; Esquena, J. pH-responsive chitosan nanofilms crosslinked with genipin. Colloids Surf. A Physicochem. Eng. Asp. 2021, 616, 126229. [Google Scholar] [CrossRef]
- Tomczak, N.; Vancso, G.J. Elasticity of single poly(amido amine) dendrimers. Macromol. Rapid Commun. 2007, 28, 1640–1644. [Google Scholar] [CrossRef]
- Kim, B.-S.; Lebedeva, O.V.; Kim, D.H.; Caminade, A.-M.; Majoral, J.-P.; Knoll, W.; Vinogradova, O.I. Assembly and mechanical properties of phosphorus dendrimer/polyelectrolyte multilayer microcapsules. Langmuir 2005, 21, 7200–7206. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Cai, Q.; Xu, F. Nanoscale mechanical properties and indentation recovery of PI@GO composites measured using AFM. Polymers 2018, 13, 1020. [Google Scholar] [CrossRef] [Green Version]
- Morozov, I.A.; Kamenetskikh, A.S.; Beliaev, A.Y.; Scherban, M.G.; Kiselkov, D.M. Low energy implantation of carbon into elastic polyurethane. Coatings 2020, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Gahlen, P.; Fröbel, S.; Karbach, A.; Gabriel, D.; Stommel, M. Experimental multi-scale approach to determine the local mechanical properties of foam base material in polyisocyanurate metal panels. Polym. Test. 2021, 93, 1069605. [Google Scholar] [CrossRef]
- Chabi, S.; Dikin, D.A.; Yin, J.; Percec, S.; Ren, F. Structure-mechanical property relations of skin-core regions of poly(p-phenylene terephthalamide) single fiber. Sci. Rep. 2019, 9, 740. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Ma, J.; Wang, C.; Li, H.; Wu, C.; Zhang, W. Precise determination of elastic modulus of cell using conical AFM probe. J. Biomech. 2021, 118, 110277. [Google Scholar] [CrossRef]
- Dias, F.V.; Serrazina, S.; Vitorino, M.; Marchese, D.; Heilmann, I.; Godinho, M.; Rodrigues, M.; Malhó, R. A role for diacylglycerol kinase 4 in signalling crosstalk during Arabidopsis pollen tuve growth. New Phytol. 2019, 222, 1434–1446. [Google Scholar] [CrossRef]
- de Araujo Dorneles, M.L.; Cardoso-Lima, R.; Noronha Souza, P.F.; Santoro Rosa, D.; Monteiro Magne, T.; Santos-Oliveira, R.; Rebelo Alencar, L.M. Zika virus (ZIKV): A new perspective on the nanomechanical and structural properties. Viruses 2022, 14, 1727. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Hirate, T.; Uehara, C.; Maruyama, H.; Uozumi, N.; Arai, F. Evaluating young’s modulus of single yeast cells based on compression using an atomic force microscope with a flat tip. Microsc. Microanal. 2021, 27, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.D.; Yin, H.; Landick, R.; Gelles, J.; Block, S.M. Stretching DNA with optical tweezers. Biophys. J. 1997, 72, 1335–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenner, J.R.; Williams, M.C.; Rouzina, I.; Bloomfield, V.A. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002, 82, 3160–3169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustamante, C.; Marko, J.F.; Siggia, E.D.; Smith, S. Entropic elasticity of lambda-phage DNA. Science 1994, 265, 1599–1600. [Google Scholar] [CrossRef] [Green Version]
- Marko, J.F.; Siggia, E.D. Stretching DNA. Macromolecules 1995, 28, 8759–8770. [Google Scholar] [CrossRef]
- Bustamante, C.; Smith, S.B.; Liphardt, J.; Smith, D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000, 10, 279–285. [Google Scholar] [CrossRef]
- Baumann, C.G.; Smith, S.B.; Bloomfield, V.A.; Bustamante, C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA 1997, 94, 6185–6190. [Google Scholar] [CrossRef] [Green Version]
- Podgornik, R.; Hansen, P.L.; Parsegian, V.A. Elastic moduli renormalization in self-interaction stretchable polyelectrolytes. J. Chem. Phys. 2000, 113, 9343. [Google Scholar] [CrossRef]
- Dao, M.; Lim, C.T.; Suresh, S. Mechanics of the human red blood cell deformed by optical tweezers. J. Mech. Phys. Solids 2003, 51, 2259–2280. [Google Scholar] [CrossRef]
- Agrawal, R.; Smart, T.; Nobre-Cardoso, J.; Richards, C.; Bhatnagar, R.; Tufail, A.; Shima, D.; Jones, P.H.; Pavesio, C. Assessment of red blood cell deformability in type 2 diabetes mellitus and diabetic retinopathy by dual optical tweezers stretching technique. Sci. Rep. 2016, 6, 15873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooke, B.M.; Mohandas, N.; Coppel, R.L. The malaria-infected red blood cell: Structural and functional changes. Adv. Parasitol. 2001, 50, 1–86. [Google Scholar] [CrossRef] [PubMed]
- Gieseler, J.; Gomez-Solano, J.R.; Magazzù, A.; Castillo, I.P.; García, L.P.; Gironella-Torrent, M.; Viader-Godoy, X.; Ritort, F.; Pesce, G.; Arzola, A.V.; et al. Optical tweezers—From calibration to applications: A tutorial. Adv. Opt. Photonics 2021, 13, 74–241. [Google Scholar] [CrossRef]
- Guck, J.; Ananthakrishnan, R.; Mahmood, H.; Moon, T.J.; Cunningham, C.C.; Käs, J. The optical stretcher: A novel laser tool to micromanipulate cells. Biophys. J. 2001, 81, 767–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irrera, A.; Magazzù, A.; Artoni, P.; Simpson, S.H.; Hanna, S.; Jones, P.H.; Priolo, F.; Gucciardi, P.G.; Maragò, O.M. Photonic torque microscopy of the nonconservative force field for optically trapped silicon nanowires. Nano Lett. 2016, 16, 4181–4188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, M.G.; Hernandez, J.; Mazzulla, A.; Provenzano, C.; Saija, R.; Sayed, R.; Vasi, S.; Magazzù, A.; Pagliusi, P.; Bartolino, R.; et al. Polarization-dependent optomechanics mediated by chiral microresonators. Nat. Commun. 2014, 5, 3656. [Google Scholar] [CrossRef] [Green Version]
- Donato, M.G.; Mazzulla, A.; Pagliusi, P.; Magazzù, A.; Hernandez, R.J.; Provenzano, C.; Gucciardi, P.G.; Maragò, O.M.; Cipparrone, G. Light-induced rotations of chiral birefringent microparticles in optical tweezers. Sci. Rep. 2016, 6, 31977. [Google Scholar] [CrossRef]
- Magazzù, A.; Ciriza, D.B.; Musolino, A.; Saidi, A.; Polimeno, P.; Donato, M.G.; Foti, A.; Gucciardi, P.G.; Iatì, M.A.; Saija, R. Investigation of dust grains by optical tweezers for space applications. Astrophys. J. 2023, 942, 1. [Google Scholar] [CrossRef]
- Watson, M.L.; Brown, D.L.; Stilgoe, A.B.; Stow, J.L.; Rubinsztein-Dunlop, H. Rotational optical tweezers for active microrheometry within living cells. Optica 2022, 9, 1066–1072. [Google Scholar] [CrossRef]
- Bishop, A.I.; Nieminen, T.A.; Heckenberg, N.; Rubinsztein-Dunlop, H. Optical microrheology using rotating laser-trapped particles. Phys. Rev. Lett. 2004, 92, 198104. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, A.S.; Nikolaev, K.G.; Novikov, A.S.; Yurchenko, S.O.; Novoselov, K.S.; Andreeva, D.V.; Skorb, E.V. Programmable soft-matter electronics. J. Phys. Chem. Lett. 2021, 12, 2017–2022. [Google Scholar] [CrossRef]
- Meng, J.-F.; Song, B.-Y.; Li, F.; Li, T.-H. Ce-MOF-based superhydrophobic polyurethane sponge reinforced by cellulose for efficient oil-water separation. Mater. Today Chem. 2023, 28, 101371. [Google Scholar] [CrossRef]
- Alamán-Díez, P.; García-Gareta, E.; Arruebo, M.; Ángeles Pérez, M. A bone-on-a-chip collagen hydrogel-based model using pre-differentiated adipose-derived stem cells for personalized bone tissue engineering. J. Biomed. Mater. Res. A. 2023, 111, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Luo, X.; Zhao, H.; Qiao, C.; Li, J.; Yi, J.; Yang, L.; Oropeza, F.J.; Hu, T.S.; Xu, Q.; et al. Recent advances in biomimetic soft robotics: Fabrication approaches, driven strategies and applications. Soft Matter 2022, 18, 7699–7734. [Google Scholar] [CrossRef] [PubMed]
- Battistella, C.; Liang, Y.; Gianneschi, N.C. Innovations in disease state responsive soft materials for targeting extracellular stimuli associated with cancer, cardiovascular disease, diabetes, and beyond. Adv. Mater. 2021, 33, e2007504. [Google Scholar] [CrossRef]
- Marcuello, C. Current and future perspectives of atomic force microscopy to elicit the intrinsic properties of soft matter at the single molecule level. AIMS Bioeng. 2022, 9, 293–306. [Google Scholar] [CrossRef]
- Marcuello, C. Present and future opportunities in the use of atomic force microscopy to address the physico-chemical properties of aquatic ecosystems at the nanoscale level. Int. Aquat. Res. 2022, 14, 231–240. [Google Scholar] [CrossRef]
- Wang, N.; Barfoot, R.; Butler, M.; Durkan, C. Effect of surface treatments on the nanomechanical properties of human hair. ACS Biomater. Sci. Eng. 2018, 4, 3063–3071. [Google Scholar] [CrossRef]
- Del Valle, A.; Torra, J.; Bondia, P.; Tone, C.M.; Pedraz, P.; Vadillo-Rodriguez, V.; Flors, C. Mechanically induced bacterial death imaged in real time: A simultaneous nanoindentation and fluorescence microscopy study. ACS Appl. Mater. Interfaces 2020, 12, 31235–31241. [Google Scholar] [CrossRef]
- Becerra, N.; Salis, B.; Tedesco, M.; Flores, S.M.; Vena, P.; Raiteri, R. AFM and fluorescence microscopy of single cells with simultaneous mechanical stimulation via electrically stretchable substrates. Materials 2021, 14, 4131. [Google Scholar] [CrossRef]
- Luo, M.; Yang, W.; Cartwright, T.N.; Higgins, J.M.G.; Chen, J. Simultaneous measurement of single-cell mechanics and cell-to-materials adhesion using fluidic force microscopy. Langmuir 2022, 38, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Deliorman, M.; Janahi, F.K.; Sukumar, P.; Glia, A.; Alnemari, R.; Fadl, S.; Chen, W.; Qasaimeh, M.A. AFM-compatible microfluidic platform for affinity-based capture and nanomechanical characterization of circulating tumor cells. Microsyst. Nanoeng. 2020, 6, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-F.; O’Callahan, B.T.; Krayev, A.; El-Khoury, P. Nanoindentation-enhanced tip-enhanced Raman Spectroscopy. J. Chem. Phys. 2021, 154, 241101. [Google Scholar] [CrossRef]
- Fraulob, M.; Pang, S.; Le Cann, S.; Vayron, R.; Laurent-Brocq, M.; Todatry, S.; Soares, J.A.N.T.; Jasiuk, I.; Haïat, G. Multimodal characterization of the bone-implant interface using Raman spectroscopy and nanoindentation. Med. Eng. Phys. 2020, 84, 60–67. [Google Scholar] [CrossRef]
- Lee, S.-H. Optimal integration of wide field illumination and holographic optical tweezers for multimodal microscopy with ultimate flexibility and versatility. Opt. Express 2018, 26, 8049–8058. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, K.K. Optical tweezers for single cells. J. R. Soc. Interface 2008, 5, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Whitley, K.D.; Comstock, M.J.; Chemla, Y.R. High-resolution “Fleezers”: Dual-trap optical tweezers combined with single-molecule fluorescence detection. Methods Mol. Biol. 2017, 1486, 183–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparkes, I.; White, R.R.; Coles, B.; Botchway, S.W.; Ward, A. Using optical tweezers combined with total internal reflection microscopy to study interactions between the ER and Golgi in plant cells. Methods Mol. Biol. 2018, 1691, 167–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, A.C.; Rusciano, G.; Ciancia, R.; Martinelli, V.; Pesce, G.; Rotoli, B.; Selvaggi, L.; Sasso, A. Spectroscopical and mechanical characterization of normal and thalassemic red blood cells by Raman tweezers. Opt. Express 2008, 16, 7943–7947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillibert, R.; Magazzù, A.; Callegari, A.; Bronte-Ciriza, D.; Foti, A.; Donato, M.G.; Maragò, O.M.; Volpe, G.; de La Chapelle, M.L.; Lagarde, F.; et al. Raman tweezers for tire and road wear micro-and nanoparticles analysis. Environ. Sci. Nano 2022, 9, 145–161. [Google Scholar] [CrossRef]
- Gillibert, R.; Balakrishan, G.; Deshoules, Q.; Tardivel, M.; Magazzù, A.; Donato, M.G.; Maragò, O.M.; de La Chapelle, M.L.; Colas, F.; Lagarde, F.; et al. Raman tweezers for small microplastics and nanoplastics identification in seawater. Environ. Sci. Technol. 2019, 53, 9003–9013. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, K.; Wang, N.; Ta, K.; Liang, P.; Yin, H.; Li, B. Laser tweezers Raman spectroscopy combined with deep learning to classify marine bacteria. Talanta 2022, 244, 123383. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Steinkühler, J.; Zhao, Z.; Lipowsky, R.; Dimova, R. Mechanical tension of biomembranes can be measured by super resolution (STED) microscopy of force-induced nanotubes. Nano Lett. 2020, 20, 3185–3191. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; He, F.; Yu, Y.; Wang, Y. Application of FRET biosensors in mechanobiology and mechanopharmacological screening. Front. Bioeng. Biotecnol. 2020, 8, 595497. [Google Scholar] [CrossRef] [PubMed]
- García-Aznar, J.M.; Nasello, G.; Hervas-Raluy, S.; Pérez, M.Á.; Gómez-Benito, M.J. Multiscale modeling of bone tissue mechanobiology. Bone 2021, 151, 116032. [Google Scholar] [CrossRef]
- Sánchez, M.T.; Pérez, M.Á.; García-Aznar, J.M. The role of fluid flow on bone mechanobiology: Mathematical modeling and simulation. Comput. Geosci. 2021, 25, 823–830. [Google Scholar] [CrossRef]
- Clegg, P.S. Characterising soft matter using machine learning. Soft Matter 2021, 17, 3991–4005. [Google Scholar] [CrossRef]
- Beltrán, G.; Navajas, D.; García-Aznar, J.M. Mechanical modeling of lung alveoli: From macroscopic behavior to cell mechano-sensing at microscopic level. J. Mech. Behav. Biomed. Mater. 2022, 126, 105043. [Google Scholar] [CrossRef]







| Equation | Ultimate Tensile Strength | Yield Strength |
|---|---|---|
| Hollomon | ||
| Swift | ||
| Voce |
| Spring Constant (k) | Young’s Modulus |
|---|---|
| 0.5 N/m | 1 MPa–20 MPa |
| 5.0 N/m | 5 MPa–500 MPa |
| 40.0 N/m | 200 MPa–2 GPa |
| 200.0 N/m | 1 GPa–20 GPa |
| 350.0 N/m | 10 GPa–100 GPa |
| Soft Matter System | Sample | Conditions | Elastic Modulus | [Ref.] |
|---|---|---|---|---|
| Biopolymer | Cellulose nanocrystals (CNCs) films | 45% R.H. (L.F) 200 nN | 10.3 ± 0.9 GPa | [168] |
| Biopolymer | Lignin films | 45% R.H (L.F) 200 nN | 6.3 ± 0.4 GPa | [168] |
| Biopolymer | Oxidized lignin films | 45% R.H (L.F) 200 nN | 11.0 ± 1.6 GPa | [168] |
| Polymer | Polypropylene (PP) | 50 °C | 1.4 ± 0.1 GPa | [43] |
| Polymer | Polybutylene succinate (PBS) | 50 °C | 548.3 ± 14.2 MPa | [43] |
| Hydrogel | Acrylamide (5.5%)–Bis(*) 0.03% | Room | 2.0 ± 0.1 kPa | [169] |
| Hydrogel | Acrylamide (12.0%)–Bis(*) 0.15% | Room | 29.0 ± 6.2 kPa | [169] |
| Hydrogel | Chitosan—genipin | pH 3, 1 h react. t. | 477 MPa | [170] |
| Hydrogel | Chitosan—genipin | pH 6, 1 h react. t. | 615 MPa | [170] |
| Dendrimer | 5 poly(amido amine) (PAMAM) | Room | 700 MPa | [171] |
| Dendrimer | Polyelectrolite microcapsules | Room | 150 MPa | [172] |
| Blend | CNC:oxidized lignin | 45% R.H (L.F) 200 nN | 13.6 ± 0.6 GPa | [114] |
| Blend | Polyimide:graphene oxide | Load force (L.F) 55 µN | 6.3 ± 0.5 GPa | [173] |
| Blend | Polyurethane:carbon | 100 impulses, 0.5 keV | 75 MPa | [174] |
| Foam | Polyisocyanurate | Room. Height 30 mm | 3.4 ± 0.4 GPa | [175] |
| Liquid crystal | Poly (p-phenylene terephthalamide) | Load force 1000 µN | 5.6 GPa | [176] |
| Biological (bacteria) | Staphylococcus epidermidis | Deionized water | 1.0 ± 0.3 MPa | [162] |
| Biological (bacteria) | Staphylococcus epidermidis | 100 mM CaCl2 | 0.6 ± 0.2 MPa | [162] |
| Biological (living cell) | Human osteosarcoma | Liquid | 34.3 ± 2.4 kPa | [177] |
| Biological (living cell) | Human skin (normal) | Room L.F 2.9 µN | 401 ± 148 MPa | [166] |
| Biological (living cell) | Patient skin (benign nevus) | Room L.F 2.9 µN | 575 ± 107 MPa | [166] |
| Biological (living cell) | Patient skin (melanoma) | Room L.F 2.9 µN | 188–787 MPa | [166] |
| Biological (plant) | Pollen tube Arabidopsis thaliana | Room L.F 6.5 nN | 46 ± 12 MPa | [178] |
| Biological (tissue) | Human colon cancer | Fixed-frozen section | 115.8 kPa | [164] |
| Biological (tissue) | Mice cortical bone | PBS. L.F 0.5 nN | 0.86 kPa | [167] |
| Biological (virus) | HK97 bacteriophage | Ident. depth 5.5 nm | 400 MPa | [165] |
| Biological (virus) | HK97 bacteriophage | Ident. depth 8.5 nm | 900 MPa | [165] |
| Biological (virus) | Zika viral particles | Room L.F 6.0 nN | 234 kPa | [179] |
| Biological (yeast) | Saccharomyces cerevisiae | Room. L.F 1.0 µN | 5.1 ± 1.5 MPa | [180] |
| Buffer Composition | (nm) | (pN) |
|---|---|---|
| 10 mM Na+ (NaHPO4 buffer, pH 7.0) | 47.4 ± 1.0 | 1008 ± 38 |
| 150 mM Na+, 5 mM Mg2+ (NaHPO4 buffer, pH 7.0) | 43.1 + 1.3 | 1205 ± 87 |
| 10 mM Na+, 100, LM spermidine (NaHPO4 buffer, pH 7.0) | 38.7 ± 1.0 | 1202 ± 83 |
| 20 mM Tris, 130 mM K+, 4 mM Mg2+ (PTC buffer, pH 8.0) | 41.0 ± 0.8 | 1277 + 57 |
| Na+ Concentration (mM) | (nm) | (pN) |
|---|---|---|
| 2.6 | 68 ± 2 | 741 ± 56 |
| 10 | 67 ± 4 | 741 ± 147 |
| 25 | 58 ± 3 | 790 ± 104 |
| 53.5 | 52 ± 1 | 1078 ± 64 |
| 100 | 48 ± 2 | 884 + 116 |
| 250 | 46 ± 1 | 1038 ± 69 |
| 500 | 47 + 2 | 1049 ± 226 |
| 1000 | 46 ± 2 | 1256 ± 217 |
| Ionic Stength (mM) | (nm) | (pN) |
|---|---|---|
| 1.86 | 94.9 ± 5.9 | 649 ± 82 |
| 3.72 | 75.7 ± 2.5 | 745 ± 100 |
| 5.58 | 76.7 ± 5.4 | 476 ± 142 |
| 7.44 | 62.2 ± 3.7 | 686 ± 65 |
| 93.0 | 65.2 ± 2.7 | 452 ± 35 |
| 18.6 | 52.9 ± 9.5 | 532 ± 67 |
| 93.0 | 51.1 ± 1.8 | 1006 ± 2 |
| 186 | 52.5 ± 12.4 | 1401 ± 313 |
| 586 | 55.9 ± 3.2 | 1435 ± 160 |
| Control Group | DM Group | DR Group | |
|---|---|---|---|
| Average unstretched cell size (μm) | 8.45 ± 0.25 | 8.68 ± 0.49 | 8.82 ± 0.32 |
| Average maximal stretched cell size (μm) | 9.04 ± 0.17 | 9.23 ± 0.49 | 9.39 ± 0.26 |
| Average difference between stretched and unstretched cell size (μm) | 0.59 ± 0.19 | 0.56 ± 0.32 | 0.56 ± 0.24 |
| Deformability index | 0.0698 ± 0.024 | 0.0645 ± 0.03 | 0.0635 ± 0.029 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magazzù, A.; Marcuello, C. Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review. Nanomaterials 2023, 13, 963. https://doi.org/10.3390/nano13060963
Magazzù A, Marcuello C. Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review. Nanomaterials. 2023; 13(6):963. https://doi.org/10.3390/nano13060963
Chicago/Turabian StyleMagazzù, Alessandro, and Carlos Marcuello. 2023. "Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review" Nanomaterials 13, no. 6: 963. https://doi.org/10.3390/nano13060963