Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H
Abstract
:1. Introduction
2. Experimental
2.1. Construction of the Mutant Strain
2.2. Bacterial Growth and Reaction Center Purification
2.3. Thermal Stability, Adsorption Spectroscopy, and Redox Potential Measurements
2.4. Reaction Center Crystallization and Data Analysis
3. Results
3.1. Absorption Spectra
3.2. Thermal Stability
3.3. P/P+ Midpoint Potential
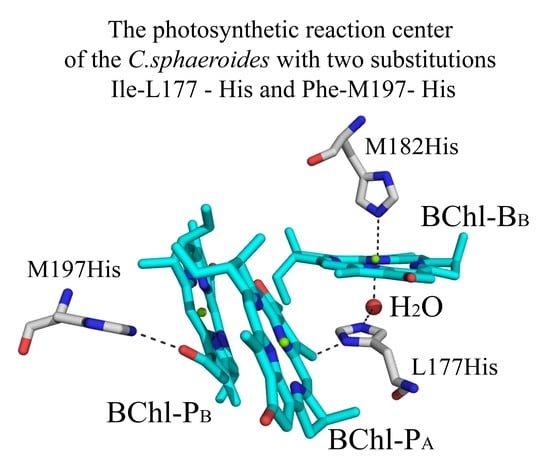
3.4. Crystal Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, J.P.; Feher, G.; Yeates, T.O.; Komiya, H.; Rees, D.C. Structure of the reaction center from Rhodobacter sphaeroides R-26: The cofactors. Proc. Natl. Acad. Sci. USA 1987, 84, 5730–5734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feher, G.; Allen, J.P.; Okamura, M.Y.; Rees, D.C. Structure and function of bacterial photosynthetic reaction centres. Nature 1989, 339, 111–116. [Google Scholar] [CrossRef]
- Yeates, T.O.; Komiya, H.; Chirino, A.; Rees, D.C.; Allen, J.P.; Feher, G. Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: Protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions. Proc. Natl. Acad. Sci. USA 1988, 85, 7993–7997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.R. Structural plasticity of reaction centers from purple bacteria. In Advances in Photosynthesis and Respiration; Hunter, C.N., Daldal, F., Thurnauer, M.C., Beatty, J.T., Eds.; Springer: Dordrecht, The Netherlands, 2009; Volume 28, pp. 295–321. [Google Scholar]
- Fufina, T.Y.; Vasilieva, L.G.; Khatypov, R.A.; Shkuropatov, A.Y.; Shuvalov, V.A. Substitution of isoleucine L177 by histidine in Rhodobacter sphaeroides reaction center results in the covalent binding of PA bacteriochlorophyll to the L subunit. FEBS Lett. 2007, 581, 5769–5773. [Google Scholar] [CrossRef] [Green Version]
- Vasilieva, L.G.; Fufina, T.Y.; Gabdulkhakov, A.G.; Leonova, M.M.; Khatypov, R.A.; Shuvalov, V.A. The site-directed mutation I(L177)H in Rhodobacter sphaeroides reaction center affects coordination of PA and BB bacteriochlorophylls. Biochim. Biophys. Acta 2012, 1817, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Fufina, T.Y.; Vasilieva, L.G.; Shuvalov, V.A. Examination of stability of mutant photosynthetic reaction center of Rhodobacter sphaeroides I(L177)H and determination of location of bacteriochlorophyll covalently bound to the protein. Biochemistry 2010, 75, 208–213. [Google Scholar] [CrossRef]
- Holden-Dye, K.; Crouch, L.I.; Williams, C.M.; Bone, R.A.; Cheng, J.; Böhles, F.; Heathcote, P.; Jones, M.R. Opposing structural changes in two symmetrical polypeptides bring about opposing changes to the thermal stability of a complex integral membrane protein. Arch. Biochem. Biophys. 2011, 505, 160–170. [Google Scholar] [CrossRef]
- Kangur, L.; Jones, M.R.; Freiberg, A. Hydrogen bonds in the vicinity of the special pair of the bacterial reaction center probed by hydrostatic high-pressure absorption spectroscopy. Biophys. Chemistr. 2017, 231, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Selikhanov, G.K.; Fufina, T.Y.; Günther, S.; Meents, A.; Gabdulkhakov, A.G.; Vasilieva, L.G. X-Ray structure of Rhodobacter sphaeroides reaction center with M197 Phe-His substitution clarifies properties of the mutant complex. IUCrJ 2022, 9, 261–271. [Google Scholar] [CrossRef]
- Khatypov, R.A.; Vasilieva, L.G.; Bolgarina, T.I.; Shuvalov, V.A. Substitution of isoleucine L177 by histidine affects the pigment composition and properties of the reaction center of the purple bacterium Rhodobacter sphaeroides. Biochemistry 2005, 70, 1256–1261. [Google Scholar] [CrossRef]
- Khmelnitskiy, A.Y.; Khatypov, R.A.; Khristin, A.M.; Leonova, M.M.; Vasilieva, L.G.; Shuvalov, V.A. Charge separation in Rhodobacter sphaeroides mutant reaction centers with increased midpoint potential of the primary electron donor. Biochemistry 2013, 78, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, J.O.; Boxer, S.G. Rapid isolation of bacterial photosynthetic reaction centers with an engineered poly-histidine tag. Biochim. Biophys. Acta 1996, 1276, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.R.; Visschers, R.W.; van Grondelle, R.; Hunter, C.N. Construction and characterization of a mutant strain of Rhodobacter sphaeroides with the reaction center as the sole pigment-protein complex. Biochemistry 1992, 31, 4458–4465. [Google Scholar] [CrossRef] [PubMed]
- Fufina, T.Y.; Vasilieva, L.G. Effects of detergents and osmolytes on thermal stability of native and mutant Rhodobacter sphaeroides reaction centers. Biochemistry 2021, 86, 517–524. [Google Scholar] [CrossRef]
- Kabsch, W. XDS . Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System v2.5. Available online: https://pymol.org/2/ (accessed on 18 December 2021).
- Hoff, A.J.; Deisenhofer, J. Photophysics of photosynthesis. Structure and spectroscopy of reaction centers of purple bacteria. Phys. Rep. 1997, 287, 242–247. [Google Scholar] [CrossRef]
- Ridge, J.P.; Fyfe, P.K.; McAuley, K.E.; Van Brederode, M.; Robert, B.; Van Grondelle, R.; Isaacs, N.W.; Cogdell, R.J.; Jones, M.R. An examination of how structural changes can affect the rate of electron transfer in a mutated bacterial photoreaction centre. Biochem. J. 2000, 351, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Murchison, H.A.; Nagarajan, V.; Parson, W.W.; Allen, J.P.; Williams, J.C. Specific alteration of the oxidation potential of the electron donor in reaction centers from Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. USA 1994, 91, 10265–10269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelshtein, A.V.; Ptitsyn, O.B. Protein Physics; Knizhnyi Dom Universitet Publisher: Moscow, Russia, 2005. (In Russian) [Google Scholar]
- Chen, M.; Schliep, M.; Willows, R.D.; Cai, Z.-L.; Neilan, B.A.; Scheer, H. A Red-Shifted Chlorophyll. Science 2010, 329, 1318–1319. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, H.L.; Okamura, M.Y. The structure and function of the cytochrome c2: Reaction center electron transfer complex from Rhodobacter Sphaeroides. Photosynth. Res. 2005, 85, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, C.; Mayneord, G.E.; Brindley, A.A.; Johnson, M.P.; Hunter, C.N. Dissecting the cytochrome c2-reaction centre interaction in bacterial photosynthesis using single molecule force spectroscopy. Biochem. J. 2019, 476, 2173–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, J.P.; Chamberlain, K.D.; Williams, J.C. Identification of amino acid residues in a proton release pathway near the bacteriochlorophyll dimer in reaction centers from Rhodobacter Sphaeroides. Photosynth. Res. 2023, 155, 23–34. [Google Scholar] [CrossRef]






| Data Collection | |
|---|---|
| Diffraction source | PETRA III, beamline P11 |
| Wavelength (Å) | 1.03312 |
| Temperature (K) | 100 |
| Detector | Pilatus 6M |
| Rotation range per image (°) | 0.1 |
| Total rotation range (°) | 180 |
| Space group | P3121 |
| a, b, c (Å) | 139.3 139.3 184.6 |
| Resolution range (Å) | 50.00–2.60 (2.67–2.60) |
| Total No. of reflections | 637 789 (43 719) |
| No. of unique reflections | 64 208 (4 564) |
| Completeness (%) | 99.7 (97.2) |
| Redundancy | 9.9 (9.6) |
| 〈 I/σ(I)〉 | 19.21 (1.04) |
| Rr.i.m.‡ | 10.7 (216.8) |
| CC1/2 | 99.9 (49.0) |
| Structure solution and refinement | |
| Resolution range (Å) | 50.00–2.60 (2.63–2.60) |
| Completeness (%) | 99.9 (100.0) |
| No. of reflections, working set | 64 207 (3 995) |
| No. of reflections, test set | 5 901 (200) |
| Rcryst | 19.43 (34.92) |
| Rfree | 21.98 (38.88) |
| R.m.s. deviations | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.15 |
| Average B factors (Å2) | 64.95 |
| Protein | 64.13 |
| Ligand | 72.15 |
| Water | 59.05 |
| Calculated DPI (Å) | 0.17 |
| Maximal estimated error (Å) | 0.31 |
| Ramachandran plot | |
| Most favoured (%) | 96.57 |
| Allowed (%) | 3.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fufina, T.Y.; Selikhanov, G.K.; Gabdulkhakov, A.G.; Vasilieva, L.G. Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H. Membranes 2023, 13, 157. https://doi.org/10.3390/membranes13020157
Fufina TY, Selikhanov GK, Gabdulkhakov AG, Vasilieva LG. Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H. Membranes. 2023; 13(2):157. https://doi.org/10.3390/membranes13020157
Chicago/Turabian StyleFufina, Tatiana Yu., Georgii K. Selikhanov, Azat G. Gabdulkhakov, and Lyudmila G. Vasilieva. 2023. "Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H" Membranes 13, no. 2: 157. https://doi.org/10.3390/membranes13020157