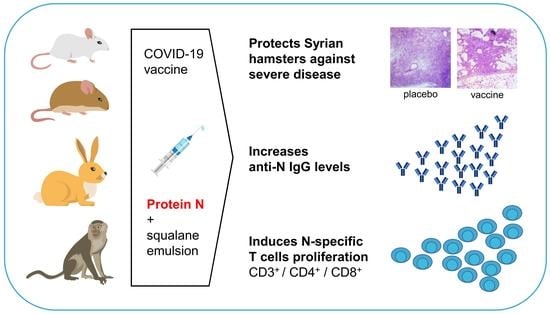
Immunogenicity and In Vivo Protective Effects of Recombinant Nucleocapsid-Based SARS-CoV-2 Vaccine Convacell®
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of N Protein and Vaccine Formulation
2.2. Stability of Vaccine
2.3. N Protein Characterization
2.4. Animals and Ethics
2.5. Safety Assessment
2.6. Measurement of N-Specific Antibodies
2.7. Measurement of N-Specific Cellular Responses
2.8. SARS-CoV-2 Infection Challenge of Syrian Hamsters
2.9. Statistical Analysis
3. Results
3.1. Production, Formulation and Biochemical Characterization of a Subunit Vaccine Based on Recombinant N, Termed Convacell®
3.2. Convacell® Induces a Robust and Long-Lasting Antibody Response in Mice, Rabbits and Syrian Hamsters
3.3. Convacell® Induced N-Specific T Cell Response with Mixed Th1/Th2 Phenotype
3.4. Vaccination with Convacell® Is Safe and Protects Syrian Hamsters against SARS-CoV-2 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Coira, J.; Sokolowska, M. SARS-CoV-2 Candidate Vaccines—Composition, Mechanisms of Action and Stages of Clinical Development. Allergy 2021, 76, 1922–1924. [Google Scholar] [CrossRef] [PubMed]
- Bok, K.; Sitar, S.; Graham, B.S.; Mascola, J.R. Accelerated COVID-19 Vaccine Development: Milestones, Lessons, and Prospects. Immunity 2021, 54, 1636–1651. [Google Scholar] [CrossRef] [PubMed]
- Heinz, F.X.; Stiasny, K. Distinguishing Features of Current COVID-19 Vaccines: Knowns and Unknowns of Antigen Presentation and Modes of Action. NPJ Vaccines 2021, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- WHO. R&D Blueprint Team COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 2 April 2023).
- Jafari, A.; Danesh Pouya, F.; Niknam, Z.; Abdollahpour-Alitappeh, M.; Rezaei-Tavirani, M.; Rasmi, Y. Current Advances and Challenges in COVID-19 Vaccine Development: From Conventional Vaccines to next-Generation Vaccine Platforms. Mol. Biol. Rep. 2022, 49, 4943–4957. [Google Scholar] [CrossRef]
- Hartley, G.E.; Edwards, E.S.J.; O’Hehir, R.E.; van Zelm, M.C. New Insights into Human Immune Memory from SARS-CoV-2 Infection and Vaccination. Allergy 2022, 77, 3553–3566. [Google Scholar] [CrossRef]
- Gattinger, P.; Kratzer, B.; Tulaeva, I.; Niespodziana, K.; Ohradanova-Repic, A.; Gebetsberger, L.; Borochova, K.; Garner-Spitzer, E.; Trapin, D.; Hofer, G.; et al. Vaccine Based on Folded Receptor Binding Domain-PreS Fusion Protein with Potential to Induce Sterilizing Immunity to SARS-CoV-2 Variants. Allergy 2022, 77, 2431–2445. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.I.; Ghany, S.; Gilkes, T.; Umakanthan, S. Review of COVID-19 Vaccine Subtypes, Efficacy and Geographical Distributions. Postgrad. Med. J. 2022, 98, 389–394. [Google Scholar] [CrossRef]
- Ferri, C.; Ursini, F.; Gragnani, L.; Raimondo, V.; Giuggioli, D.; Foti, R.; Caminiti, M.; Olivo, D.; Cuomo, G.; Visentini, M.; et al. Impaired Immunogenicity to COVID-19 Vaccines in Autoimmune Systemic Diseases. High Prevalence of Non-Response in Different Patients’ Subgroups. J. Autoimmun. 2021, 125, 102744. [Google Scholar] [CrossRef]
- Mair, M.J.; Mitterer, M.; Gattinger, P.; Berger, J.M.; Trutschnig, W.; Bathke, A.C.; Gansterer, M.; Berghoff, A.S.; Laengle, S.; Gottmann, L.; et al. Enhanced SARS-CoV-2 Breakthrough Infections in Patients with Hematologic and Solid Cancers Due to Omicron. Cancer Cell 2022, 40, 444–446. [Google Scholar] [CrossRef]
- Wagner, A.; Garner-Spitzer, E.; Schötta, A.-M.; Orola, M.; Wessely, A.; Zwazl, I.; Ohradanova-Repic, A.; Weseslindtner, L.; Tajti, G.; Gebetsberger, L.; et al. SARS-CoV-2-MRNA Booster Vaccination Reverses Non-Responsiveness and Early Antibody Waning in Immunocompromised Patients—A Phase Four Study Comparing Immune Responses in Patients with Solid Cancers, Multiple Myeloma and Inflammatory Bowel Disease. Front. Immunol. 2022, 13, 889138. [Google Scholar] [CrossRef]
- Tea, F.; Ospina Stella, A.; Aggarwal, A.; Ross Darley, D.; Pilli, D.; Vitale, D.; Merheb, V.; Lee, F.X.Z.; Cunningham, P.; Walker, G.J.; et al. SARS-CoV-2 Neutralizing Antibodies: Longevity, Breadth, and Evasion by Emerging Viral Variants. PLoS Med. 2021, 18, e1003656. [Google Scholar] [CrossRef]
- Bonnet, B.; Chabrolles, H.; Archimbaud, C.; Brebion, A.; Cosme, J.; Dutheil, F.; Lambert, C.; Junda, M.; Mirand, A.; Ollier, A.; et al. Decline of Humoral and Cellular Immune Responses Against SARS-CoV-2 6 Months After Full BNT162b2 Vaccination in Hospital Healthcare Workers. Front. Immunol. 2022, 13, 842912. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, G.; Wang, Y.; Zhang, Q.; Ren, L.; Gu, X.; Huang, T.; Zhong, J.; Wang, Y.; Wang, X.; et al. SARS-CoV-2-Specific Antibody and T-Cell Responses 1 Year after Infection in People Recovered from COVID-19: A Longitudinal Cohort Study. Lancet Microbe 2022, 3, e348–e356. [Google Scholar] [CrossRef] [PubMed]
- Barda, N.; Dagan, N.; Ben-Shlomo, Y.; Kepten, E.; Waxman, J.; Ohana, R.; Hernán, M.A.; Lipsitch, M.; Kohane, I.; Netzer, D.; et al. Safety of the BNT162b2 MRNA COVID-19 Vaccine in a Nationwide Setting. N. Engl. J. Med. 2021, 385, 1078–1090. [Google Scholar] [CrossRef]
- Dionne, A.; Sperotto, F.; Chamberlain, S.; Baker, A.L.; Powell, A.J.; Prakash, A.; Castellanos, D.A.; Saleeb, S.F.; de Ferranti, S.D.; Newburger, J.W.; et al. Association of Myocarditis with BNT162b2 Messenger RNA COVID-19 Vaccine in a Case Series of Children. JAMA Cardiol. 2021, 6, 1446–1450. [Google Scholar] [CrossRef]
- Foltran, D.; Delmas, C.; Flumian, C.; De Paoli, P.; Salvo, F.; Gautier, S.; Drici, M.-D.; Karsenty, C.; Montastruc, F. Myocarditis and Pericarditis in Adolescents after First and Second Doses of MRNA COVID-19 Vaccines. Eur. Heart J. Qual. Care Clin. Outcomes 2022, 8, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 NCov-19 Vaccination. N. Engl. J. Med. 2021, 384, 2092–2101. [Google Scholar] [CrossRef] [PubMed]
- Scully, M.; Singh, D.; Lown, R.; Poles, A.; Solomon, T.; Levi, M.; Goldblatt, D.; Kotoucek, P.; Thomas, W.; Lester, W. Pathologic Antibodies to Platelet Factor 4 after ChAdOx1 NCoV-19 Vaccination. N. Engl. J. Med. 2021, 384, 2202–2211. [Google Scholar] [CrossRef]
- Pavord, S.; Scully, M.; Hunt, B.J.; Lester, W.; Bagot, C.; Craven, B.; Rampotas, A.; Ambler, G.; Makris, M. Clinical Features of Vaccine-Induced Immune Thrombocytopenia and Thrombosis. N. Engl. J. Med. 2021, 385, 1680–1689. [Google Scholar] [CrossRef]
- Cabanillas, B.; Akdis, C.A.; Novak, N. Allergic Reactions to the First COVID-19 Vaccine: A Potential Role of Polyethylene Glycol? Allergy 2021, 76, 1617–1618. [Google Scholar] [CrossRef]
- Cabanillas, B.; Novak, N.; Akdis, C.A. The Form of PEG Matters: PEG Conjugated with Lipids and Not PEG Alone Could Be the Specific Form Involved in Allergic Reactions to COVID-19 Vaccines. Allergy 2022, 77, 1658–1660. [Google Scholar] [CrossRef]
- Patone, M.; Handunnetthi, L.; Saatci, D.; Pan, J.; Katikireddi, S.V.; Razvi, S.; Hunt, D.; Mei, X.W.; Dixon, S.; Zaccardi, F.; et al. Neurological Complications after First Dose of COVID-19 Vaccines and SARS-CoV-2 Infection. Nat. Med. 2021, 27, 2144–2153. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Salerno, M.; Esposito, M.; Di Nunno, N.; Zamboni, P.; Pomara, C. Autopsy Findings and Causality Relationship between Death and COVID-19 Vaccination: A Systematic Review. J. Clin. Med. 2021, 10, 5876. [Google Scholar] [CrossRef] [PubMed]
- Yonker, L.M.; Swank, Z.; Bartsch, Y.C.; Burns, M.D.; Kane, A.; Boribong, B.P.; Davis, J.P.; Loiselle, M.; Novak, T.; Senussi, Y.; et al. Circulating Spike Protein Detected in Post–COVID-19 MRNA Vaccine Myocarditis. Circulation 2023, 147, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Valtanen, P.; Hope, C.M.; Masavuli, M.G.; Yeow, A.E.L.; Balachandran, H.; Mekonnen, Z.A.; Al-Delfi, Z.; Abayasingam, A.; Agapiou, D.; Stella, A.O.; et al. SARS-CoV-2 Omicron Variant Escapes Neutralizing Antibodies and T Cell Responses More Efficiently than Other Variants in Mild COVID-19 Convalescents. Cell Rep. Med. 2022, 3, 100651. [Google Scholar] [CrossRef]
- Dolton, G.; Rius, C.; Hasan, M.S.; Wall, A.; Szomolay, B.; Behiry, E.; Whalley, T.; Southgate, J.; Fuller, A.; Morin, T.; et al. Emergence of Immune Escape at Dominant SARS-CoV-2 Killer T Cell Epitope. Cell 2022, 185, 2936–2951.e19. [Google Scholar] [CrossRef]
- Gattinger, P.; Tulaeva, I.; Borochova, K.; Kratzer, B.; Trapin, D.; Kropfmüller, A.; Pickl, W.F.; Valenta, R. Omicron: A SARS-CoV-2 Variant of Real Concern. Allergy 2022, 77, 1616–1620. [Google Scholar] [CrossRef]
- Lu, L.; Mok, B.W.Y.; Chen, L.L.; Chan, J.M.C.; Tsang, O.T.Y.; Lam, B.H.S.; Chuang, V.W.M.; Chu, A.W.H.; Chan, W.M.; Ip, J.D.; et al. Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant by Sera from BNT162b2 or CoronaVac Vaccine Recipients. Clin. Infect. Dis. 2022, 75, e822–e826. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Shaw, R.H.; Supasa, P.; Liu, C.; Stuart, A.S.; Pollard, A.J.; Liu, X.; Lambe, T.; Crook, D.; Stuart, D.I.; et al. Reduced Neutralisation of SARS-CoV-2 Omicron B.1.1.529 Variant by Post-Immunisation Serum. Lancet 2022, 399, 234–236. [Google Scholar] [CrossRef]
- Yang, H.; Rao, Z. Structural Biology of SARS-CoV-2 and Implications for Therapeutic Development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef]
- Dai, L.; Gao, G.F. Viral Targets for Vaccines against COVID-19. Nat. Rev. Immunol. 2021, 21, 73–82. [Google Scholar] [CrossRef]
- Pack, S.M.; Peters, P.J. SARS-CoV-2-Specific Vaccine Candidates; the Contribution of Structural Vaccinology. Vaccines 2022, 10, 236. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Banerjee, A.; Ray, S. Development of New Vaccine Target against SARS-CoV2 Using Envelope (E) Protein: An Evolutionary, Molecular Modeling and Docking Based Study. Int. J. Biol. Macromol. 2021, 172, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The Nucleocapsid Protein of SARS–CoV-2: A Target for Vaccine Development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef]
- Phatarphekar, A.; Reddy, G.E.C.V.; Gokhale, A.; Karanam, G.; Kuchroo, P.; Shinde, K.; Masand, G.; Pagare, S.; Khadpe, N.; Pai, S.S.; et al. RelCoVax®, a Two Antigen Subunit Protein Vaccine Candidate against SARS-CoV-2 Induces Strong Immune Responses in Mice. Vaccine 2022, 40, 4522–4530. [Google Scholar] [CrossRef] [PubMed]
- Thura, M.; Sng, J.X.E.; Ang, K.H.; Li, J.; Gupta, A.; Hong, J.M.; Hong, C.W.; Zeng, Q. Targeting Intra-Viral Conserved Nucleocapsid (N) Proteins as Novel Vaccines against SARS-CoVs. Biosci. Rep. 2021, 41, BSR20211491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Guo, J.; Wan, X.; Zhou, J.-G.; Jin, W.-P.; Lu, J.; Wang, W.-H.; Yang, A.-N.; Liu, D.X.; Shi, Z.-L.; et al. Biochemical and Antigenic Characterization of the Structural Proteins and Their Post-Translational Modifications in Purified SARS-CoV-2 Virions of an Inactivated Vaccine Candidate. Emerg. Microbes Infect. 2020, 9, 2653–2662. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wei, Y.; Liang, H.; Ji, W.; Chang, Z.; Xie, S.; Wang, Y.; Li, W.; Liu, Y.; Wu, H.; et al. Comparison of Physical and Biochemical Characterizations of SARS-CoV-2 Inactivated by Different Treatments. Viruses 2022, 14, 1938. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-Specific T Cell Immunity in Cases of COVID-19 and SARS, and Uninfected Controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- López-Muñoz, A.D.; Kosik, I.; Holly, J.; Yewdell, J.W. Cell Surface SARS-CoV-2 Nucleocapsid Protein Modulates Innate and Adaptive Immunity. Sci. Adv. 2022, 8, eabp9770. [Google Scholar] [CrossRef]
- Hong, S.-H.; Oh, H.; Park, Y.W.; Kwak, H.W.; Oh, E.Y.; Park, H.-J.; Kang, K.W.; Kim, G.; Koo, B.-S.; Hwang, E.-H.; et al. Immunization with RBD-P2 and N Protects against SARS-CoV-2 in Nonhuman Primates. Sci. Adv. 2021, 7, eabg7156. [Google Scholar] [CrossRef] [PubMed]
- Sieling, P.; King, T.; Wong, R.; Nguyen, A.; Wnuk, K.; Gabitzsch, E.; Rice, A.; Adisetiyo, H.; Hermreck, M.; Verma, M.; et al. Prime HAd5 Spike + Nucleocapsid Vaccination Induces Ten-Fold Increases in Mean T-Cell Responses in Phase 1 Subjects That Are Sustained Against Spike Variants. medRxiv 2021. [Google Scholar] [CrossRef]
- Brentville, V.; Vankemmelbeke, M.; Metheringham, R.; Symonds, P.; Cook, K.; Urbanowicz, R.; Tsoleridis, T.; Coleman, C.; Chang, K.-C.; Skinner, A.; et al. A Novel Bivalent DNA Vaccine Encoding Both Spike Protein Receptor-Binding Domain and Nucleocapsid Protein of SARS-CoV-2 to Elicit T Cell and Neutralising Antibody Responses That Cross React with Variants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kuwentrai, C.; Yu, J.; Zhang, B.; Hu, Y.; Dou, Y.; Gong, H.; Huang, J.-D.; Xu, C. Induction of Humoral and Cellular Immunity by Intradermal Delivery of SARS-CoV-2 Nucleocapsid Protein Using Dissolvable Microneedles. J. Immunol. Res. 2021, 2021, 5531220. [Google Scholar] [CrossRef]
- Matchett, W.E.; Joag, V.; Stolley, J.M.; Shepherd, F.K.; Quarnstrom, C.F.; Mickelson, C.K.; Wijeyesinghe, S.; Soerens, A.G.; Becker, S.; Thiede, J.M.; et al. Cutting Edge: Nucleocapsid Vaccine Elicits Spike-Independent SARS-CoV-2 Protective Immunity. J. Immunol. 2021, 207, 376–379. [Google Scholar] [CrossRef]
- He, J.; Huang, J.; Zhang, Y.; Zhang, J. SARS-CoV-2 Nucleocapsid Protein Intranasal Inoculation Induces Local and Systemic T Cell Responses in Mice. J. Med. Virol. 2021, 93, 1923–1925. [Google Scholar] [CrossRef]
- Studier, F.W. Protein Production by Auto-Induction in High-Density Shaking Cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets: Procedure and Some Applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Bairoch, A. The SWISS-PROT Protein Sequence Database and Its Supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Russian State Pharmacopoeia 14th ed. eBook. Moscow; 2018. Chapter GPA.1.2.4.0005.15, Pyrogenicity. Available online: https://femb.ru/record/pharmacopea14 (accessed on 4 January 2023).
- Kozlovskaya, L.; Piniaeva, A.; Ignatyev, G.; Selivanov, A.; Shishova, A.; Kovpak, A.; Gordeychuk, I.; Ivin, Y.; Berestovskaya, A.; Prokhortchouk, E.; et al. Isolation and Phylogenetic Analysis of SARS-CoV-2 Variants Collected in Russia during the COVID-19 Outbreak. Int. J. Infect. Dis. 2020, 99, 40–46. [Google Scholar] [CrossRef]
- Osterrieder, N.; Bertzbach, L.D.; Dietert, K.; Abdelgawad, A.; Vladimirova, D.; Kunec, D.; Hoffmann, D.; Beer, M.; Gruber, A.D.; Trimpert, J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses 2020, 12, 779. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of Two Random Variables Is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Smits, V.A.J.; Hernández-Carralero, E.; Paz-Cabrera, M.C.; Cabrera, E.; Hernández-Reyes, Y.; Hernández-Fernaud, J.R.; Gillespie, D.A.; Salido, E.; Hernández-Porto, M.; Freire, R. The Nucleocapsid Protein Triggers the Main Humoral Immune Response in COVID-19 Patients. Biochem. Biophys. Res. Commun. 2021, 543, 45–49. [Google Scholar] [CrossRef]
- Van Elslande, J.; Oyaert, M.; Ailliet, S.; Van Ranst, M.; Lorent, N.; Vande Weygaerde, Y.; André, E.; Lagrou, K.; Vandendriessche, S.; Vermeersch, P. Longitudinal Follow-up of IgG Anti-Nucleocapsid Antibodies in SARS-CoV-2 Infected Patients up to Eight Months after Infection. J. Clin. Virol. 2021, 136, 104765. [Google Scholar] [CrossRef] [PubMed]
- St. Petersburg Research Institute of Vaccines and Sera. Study of the Immunogenicity, Safety and Tolerability of the Convacell Vaccine. Available online: https://clinicaltrials.gov/ct2/show/NCT05156723 (accessed on 2 April 2023).
- Lyons, A.B.; Blake, S.J.; Doherty, K.V. Flow Cytometric Analysis of Cell Division by Dilution of CFSE and Related Dyes. Curr. Protoc. Cytom. 2013, 64, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C. Squalene and Squalane Emulsions as Adjvants. Methods 1999, 19, 87–93. [Google Scholar] [CrossRef]
- Fielding, C.A.; Sabberwal, P.; Williamson, J.C.; Greenwood, E.J.; Crozier, T.W.; Zelek, W.; Seow, J.; Graham, C.; Huettner, I.; Edgeworth, J.D.; et al. SARS-CoV-2 Host-Shutoff Impacts Innate NK Cell Functions, but Antibody-Dependent NK Activity Is Strongly Activated through Non-Spike Antibodies. eLife 2022, 11, e74489. [Google Scholar] [CrossRef]
- Jia, Q.; Bielefeldt-Ohmann, H.; Maison, R.M.; Masleša-Galić, S.; Cooper, S.K.; Bowen, R.A.; Horwitz, M.A. Replicating Bacterium-Vectored Vaccine Expressing SARS-CoV-2 Membrane and Nucleocapsid Proteins Protects against Severe COVID-19-like Disease in Hamsters. NPJ Vaccines 2021, 6, 47. [Google Scholar] [CrossRef]
- Wraith, D.C.; Vessey, A.E.; Askonas, B.A. Purified Influenza Virus Nucleoprotein Protects Mice from Lethal Infection. J. Gen. Virol. 1987, 68, 433–440. [Google Scholar] [CrossRef]
- Huang, B.; Wang, W.; Li, R.; Wang, X.; Jiang, T.; Qi, X.; Gao, Y.; Tan, W.; Ruan, L. Influenza A Virus Nucleoprotein Derived from Escherichia Coli or Recombinant Vaccinia (Tiantan) Virus Elicits Robust Cross-Protection in Mice. Virol. J. 2012, 9, 322. [Google Scholar] [CrossRef] [PubMed]
- Lazo, L.; Valdes, I.; Guillén, G.; Hermida, L.; Gil, L. Aiming at the Heart: The Capsid Protein of Dengue Virus as a Vaccine Candidate. Expert Rev. Vaccines 2019, 18, 161–173. [Google Scholar] [CrossRef]
- Kratzer, B.; Schlax, L.C.; Gattinger, P.; Waidhofer-Söllner, P.; Trapin, D.; Tauber, P.A.; Sehgal, A.N.A.; Körmöczi, U.; Rottal, A.; Feichter, M.; et al. Combined Assessment of S- and N-specific IL-2 and IL-13 Secretion and CD69 Neo-expression for Discrimination of Post–Infection and Post-vaccination Cellular SARS-CoV-2-specific Immune Response. Allergy 2022, 77, 3408–3425. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Frank, E.; Gerhard, W. Expression of Influenza A Virus Internal Antigens on the Surface of Infected P815 Cells. J. Immunol. 1981, 126, 1814–1819. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Bennink, J.R.; Mackett, M.; Lefrancois, L.; Lyles, D.S.; Moss, B. Recognition of Cloned Vesicular Stomatitis Virus Internal and External Gene Products by Cytotoxic T Lymphocytes. J. Exp. Med. 1986, 163, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Marie, J.C.; Saltel, F.; Escola, J.-M.; Jurdic, P.; Wild, T.F.; Horvat, B. Cell Surface Delivery of the Measles Virus Nucleoprotein: A Viral Strategy To Induce Immunosuppression. J. Virol. 2004, 78, 11952–11961. [Google Scholar] [CrossRef]
- Céspedes, P.F.; Bueno, S.M.; Ramírez, B.A.; Gomez, R.S.; Riquelme, S.A.; Palavecino, C.E.; Mackern-Oberti, J.P.; Mora, J.E.; Depoil, D.; Sacristán, C.; et al. Surface Expression of the HRSV Nucleoprotein Impairs Immunological Synapse Formation with T Cells. Proc. Natl. Acad. Sci. USA 2014, 111, E3214–E3223. [Google Scholar] [CrossRef] [PubMed]
- Straub, T.; Schweier, O.; Bruns, M.; Nimmerjahn, F.; Waisman, A.; Pircher, H. Nucleoprotein-Specific Nonneutralizing Antibodies Speed up LCMV Elimination Independently of Complement and FcγR: Immunity to Infection. Eur. J. Immunol. 2013, 43, 2338–2348. [Google Scholar] [CrossRef] [PubMed]
- Ikuta, K.; Morita, C.; Miyake, S.; Ito, T.; Okabayashi, M.; Sano, K.; Nakai, M.; Hirai, K.; Kato, S. Expression of Human Immunodeficiency Virus Type 1 (HIV-1) Gag Antigens on the Surface of a Cell Line Persistently Infected with HIV-1 That Highly Expresses HIV-1 Antigens. Virology 1989, 170, 408–417. [Google Scholar] [CrossRef]
- Rhodes, D.A.; Isenberg, D.A. TRIM21 and the Function of Antibodies inside Cells. Trends Immunol. 2017, 38, 916–926. [Google Scholar] [CrossRef]
- McEwan, W.A.; Mallery, D.L.; Rhodes, D.A.; Trowsdale, J.; James, L.C. Intracellular Antibody-Mediated Immunity and the Role of TRIM21: Insights & Perspectives. BioEssays 2011, 33, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Savastano, A.; Ibáñez de Opakua, A.; Rankovic, M.; Zweckstetter, M. Nucleocapsid Protein of SARS-CoV-2 Phase Separates into RNA-Rich Polymerase-Containing Condensates. Nat. Commun. 2020, 11, 6041. [Google Scholar] [CrossRef] [PubMed]
- Tilocca, B.; Soggiu, A.; Sanguinetti, M.; Musella, V.; Britti, D.; Bonizzi, L.; Urbani, A.; Roncada, P. Comparative Computational Analysis of SARS-CoV-2 Nucleocapsid Protein Epitopes in Taxonomically Related Coronaviruses. Microbes Infect. 2020, 22, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Rabdano, S.; Mukhin, V.; Makarov, V.; Rudakov, G.; Ruzanova, E.; Arakelov, S.; Khaitov, M.; Yudin, S.; Kryuchko, D.; Berzin, I.; et al. N protein based vaccine against SARS-CoV-2 produces a strong T cell immune response to N Protein of novel strains. Med. Extreme Situat. 2022, 3, 5–12. [Google Scholar] [CrossRef]





| Animal, Sex | Cell Phenotype | Days after First Immunization | |||
|---|---|---|---|---|---|
| 0 | 14 | 28 | 35 | ||
| 1, F | CD3+ | - | 0.82 | - | - |
| CD3+CD4+CD8− | - | 0.63 | - | - | |
| CD3+CD4−CD8+ | - | 0.54 | 0.55 | - | |
| 2, M | CD3+ | - | - | 0.57 | - |
| CD3+CD4+CD8− | - | - | - | - | |
| CD3+CD4−CD8+ | - | - | - | - | |
| 3, M | CD3+ | - | 0.76 | 0.90 | - |
| CD3+CD4+CD8− | - | 1.07 | 0.81 | - | |
| CD3+CD4−CD8+ | - | - | 0.52 | - | |
| 4, F | CD3+ | - | 0.71 | - | - |
| CD3+CD4+CD8− | - | - | - | - | |
| CD3+CD4−CD8+ | - | 1.09 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabdano, S.O.; Ruzanova, E.A.; Pletyukhina, I.V.; Saveliev, N.S.; Kryshen, K.L.; Katelnikova, A.E.; Beltyukov, P.P.; Fakhretdinova, L.N.; Safi, A.S.; Rudakov, G.O.; et al. Immunogenicity and In Vivo Protective Effects of Recombinant Nucleocapsid-Based SARS-CoV-2 Vaccine Convacell®. Vaccines 2023, 11, 874. https://doi.org/10.3390/vaccines11040874
Rabdano SO, Ruzanova EA, Pletyukhina IV, Saveliev NS, Kryshen KL, Katelnikova AE, Beltyukov PP, Fakhretdinova LN, Safi AS, Rudakov GO, et al. Immunogenicity and In Vivo Protective Effects of Recombinant Nucleocapsid-Based SARS-CoV-2 Vaccine Convacell®. Vaccines. 2023; 11(4):874. https://doi.org/10.3390/vaccines11040874
Chicago/Turabian StyleRabdano, Sevastyan O., Ellina A. Ruzanova, Iuliia V. Pletyukhina, Nikita S. Saveliev, Kirill L. Kryshen, Anastasiia E. Katelnikova, Petr P. Beltyukov, Liliya N. Fakhretdinova, Ariana S. Safi, German O. Rudakov, and et al. 2023. "Immunogenicity and In Vivo Protective Effects of Recombinant Nucleocapsid-Based SARS-CoV-2 Vaccine Convacell®" Vaccines 11, no. 4: 874. https://doi.org/10.3390/vaccines11040874