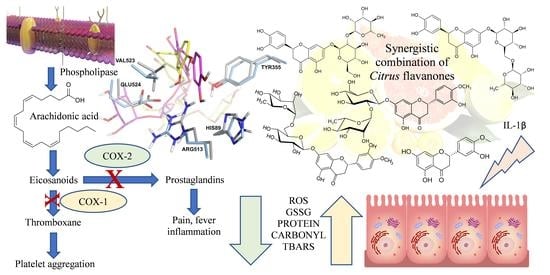
Synergistic Combination of Citrus Flavanones as Strong Antioxidant and COX-Inhibitor Agent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Test Solutions Preparation
2.3. Inhibitory Activity on Human COX-1 and COX-2
2.4. Molecular Modeling Studies
2.5. Cell-Based Assays
2.5.1. Cell Model
2.5.2. Cell Treatment
2.5.3. Detection of PGE2
2.5.4. Detection of Oxidative Stress Markers
2.5.5. Post-Quality Control Assays
2.6. Evaluation of the Synergistic Effect by CompuSyn Software
2.7. Statistical Analysis
3. Results
3.1. In Vitro Inhibitory Activity on Human COX-1 and COX-2
3.2. Molecular Modeling Studies
3.3. Anti-Inflammatory Activity
3.4. Antioxidant Activity
3.5. Post-Quality Control Assays
3.6. Evaluation of the Synergistic Effects
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barreca, D.; Gattuso, G.; Bellocco, E.; Calderaro, A.; Trombetta, D.; Smeriglio, A.; Laganà, G.; Daglia, M.; Meneghini, S.; Nabavi, S.M. Flavanones: Citrus phytochemical with health-promoting properties. Biofactors 2017, 43, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Barreca, D.; Mandalari, G.; Calderaro, A.; Smeriglio, A.; Trombetta, D.; Felice, M.R.; Gattuso, G. Citrus flavones: An update on sources, biological functions, and health promoting properties. Plants 2020, 9, 288. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Ikram, M.; Hahm, J.R.; Kim, M.O. Antioxidant and anti-inflammatory effects of citrus flavonoid hesperetin: Special focus on neurological disorders. Antioxidants 2020, 9, 609. [Google Scholar] [CrossRef] [PubMed]
- Bagetta, D.; Maruca, A.; Lupia, A.; Mesiti, F.; Catalano, R.; Romeo, I.; Moraca, F.; Ambrosio, F.A.; Costa, G.; Artese, A.; et al. Mediterranean products as promising source of multi-target agents in the treatment of metabolic syndrome. Eur. J. Med. Chem. 2020, 186, 111903. [Google Scholar] [CrossRef] [PubMed]
- Rees, A.; Dodd, G.F.; Spencer, J.P.E. The effects of flavonoids on cardiovascular health: A review of human intervention trials and implications for cerebrovascular function. Nutrients 2018, 10, 1852. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Hernández Bautista, R.J.; Sandhu, M.A.; Hussein, O.E. Beneficial effects of Citrus flavonoids on cardiovascular and metabolic health. Oxid. Med. Cell Longev. 2019, 2019, 5484138. [Google Scholar] [CrossRef]
- Musumeci, L.; Maugeri, A.; Cirmi, S.; Lombardo, G.E.; Russo, C.; Gangemi, S.; Calapai, G.; Navarra, M. Citrus fruits and their flavonoids in inflammatory bowel disease: An overview. Nat. Prod. Res. 2020, 34, 122–136. [Google Scholar] [CrossRef]
- Stevens, Y.; Rymenant, E.V.; Grootaert, C.; Camp, J.V.; Possemiers, S.; Masclee, A.; Jonkers, D. The intestinal fate of Citrus flavanones and their effects on gastrointestinal health. Nutrients 2019, 11, 1464. [Google Scholar] [CrossRef]
- Tanveer, A.; Akram, K.; Farooq, U.; Hayat, Z.; Shafi, A. Management of diabetic complications through fruit flavonoids as a natural remedy. Crit. Rev. Food Sci. Nutr. 2017, 57, 1411–1422. [Google Scholar] [CrossRef]
- Gandhi, G.R.; Vasconcelos, A.B.S.; Wu, D.T.; Li, H.B.; Antony, P.J.; Li, H.; Geng, F.; Gurgel, R.Q.; Narain, N.; Gan, R.Y. Citrus flavonoids as promising phytochemicals targeting diabetes and related complications: A systematic review of in vitro and in vivo studies. Nutrients 2020, 12, 2907. [Google Scholar] [CrossRef]
- Koolaji, N.; Shammugasamy, B.; Schindeler, A.; Dong, Q.; Dehghani, F.; Valtchev, P. Citrus peel flavonoids as potential cancer prevention agents. Curr. Dev. Nutr. 2020, 4, nzaa025. [Google Scholar] [CrossRef] [PubMed]
- Smeriglio, A.; Marcoccia, D.; Denaro, M.; Trombetta, D. Nutraceuticals in the treatment of inflammatory bowel disease: How the panorama has changed in the last decade? Curr. Med. Chem. 2022. [Google Scholar] [CrossRef]
- Salaritabar, A.; Darvishi, B.; Hadjiakhoondi, F.; Manayi, A.; Sureda, A.; Nabavi, S.F.; Fitzpatrick, L.R.; Nabavi, S.M.; Bishayee, A. Therapeutic potential of flavonoids in inflammatory bowel disease: A comprehensive review. World J. Gastroenterol. 2017, 23, 5097–5114. [Google Scholar] [CrossRef]
- Chang, H.; Lei, L.; Zhou, Y.; Ye, F.; Zhao, G. Dietary flavonoids and the risk of colorectal cancer: An updated meta-analysis of epidemiological studies. Nutrients 2018, 10, 950. [Google Scholar] [CrossRef] [PubMed]
- Fusco, R.; Cirmi, S.; Gugliandolo, E.; Di Paola, R.; Cuzzocrea, S.; Navarra, M. A flavonoid-rich extract of orange juice reduced oxidative stress in an experimental model of inflammatory bowel disease. J. Funct. Foods 2017, 30, 168–178. [Google Scholar] [CrossRef]
- Gholap, P.A.; Nirmal, S.A.; Pattan, S.R.; Pal, S.C.; Mandal, S.C. Potential of Moringa oleifera root and Citrus sinensis fruit rind extracts in the treatment of ulcerative colitis in mice. Pharm. Biol. 2012, 50, 1297–1302. [Google Scholar] [CrossRef]
- Khan, R.A.; Mallick, N.; Feroz, Z. Anti-inflammatory effects of Citrus sinensis L., Citrus paradisi L. and their combinations. Pak. J. Pharm. Sci. 2016, 29, 843–852. [Google Scholar]
- He, W.; Li, Y.; Liu, M.; Yu, H.; Chen, Q.; Chen, Y.; Ruan, J.; Ding, Z.; Zhang, Y.; Wang, T. Citrus aurantium L. and its flavonoids regulate TNBS-induced inflammatory bowel disease through anti-inflammation and suppressing isolated jejunum contraction. Int. J. Mol. Sci. 2018, 19, 3057. [Google Scholar] [CrossRef]
- Abe, H.; Ishioka, M.; Fujita, Y.; Umeno, A.; Yasunaga, M.; Sato, A.; Ohnishi, S.; Suzuki, S.; Ishida, N.; Shichiri, M.; et al. Yuzu (Citrus junos Tanaka) peel attenuates dextran sulfate sodium-induced murine experimental colitis. J. Oleo. Sci. 2018, 67, 335–344. [Google Scholar] [CrossRef]
- Denaro, M.; Smeriglio, A.; Trombetta, D. Antioxidant and anti-inflammatory activity of citrus flavanones mix and its stability after in vitro simulated digestion. Antioxidants 2021, 10, 140. [Google Scholar] [CrossRef]
- Kamiloglu, S.; Capanoglu, E.; Bilen, F.D.; Gonzales, G.B.; Grootaert, C.; Van de Wiele, T.; Van Camp, J. Bioaccessibility of polyphenols from plant-processing byproducts of black carrot (Daucus carota L.). J. Agric. Food Chem. 2016, 64, 2450–2458. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Uddin, M.J.; Banerjee, S.; Duggan, K.; Musee, J.; Kiefer, J.R.; Ghebreselasie, K.; Rouzer, C.A.; Marnett, L.J. Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site. J. Biol. Chem. 2019, 294, 8690–8698. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Alhindi, T.; Zhang, Z.; Ruelens, P.; Coenen, H.; Degroote, H.; Iraci, N.; Geuten, K. Protein interaction evolution from promiscuity to specificity with reduced flexibility in an increasingly complex network. Sci. Rep. 2017, 7, 44948. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Carbone, D.; Vestuto, V.; Ferraro, M.R.; Ciaglia, T.; Pecoraro, C.; Sommella, E.; Cascioferro, S.; Salviati, E.; Novi, S.; Tecce, M.F.; et al. Metabolomics-assisted discovery of a new anticancer GLS-1 inhibitor chemotype from a nortopsentin-inspired library: From phenotype screening to target identification. Eur. J. Med. Chem. 2022, 234, 114233. [Google Scholar] [CrossRef] [PubMed]
- Denaro, M.; Smeriglio, A.; De Francesco, C.; Xiao, J.; Cornara, L.; Trombetta, D. In vitro intestinal transport and anti-inflammatory properties of ideain across Caco-2 transwell model. Fitoterapia 2020, 146, 104723. [Google Scholar] [CrossRef]
- Tesoriere, L.; Attanzio, A.; Allegra, M.; Gentile, C.; Livrea, M.A. Indicaxanthin inhibits NADPH oxidase (NOX)-1 activation and NF-κB-dependent release of inflammatory mediators and prevents the increase of epithelial permeability in IL-1β-exposed Caco-2 cells. Br. J. Nutr. 2014, 111, 415–423. [Google Scholar] [CrossRef]
- Smeriglio, A.; De Francesco, C.; Denaro, M.; Trombetta, D. Prickly pear betalain-rich extracts as new promising strategy for intestinal inflammation: Plant complex vs. main isolated bioactive compounds. Front. Pharmacol. 2021, 12, 722398. [Google Scholar] [CrossRef]
- Nobili, V.; Alisi, A.; Mosca, A.; Crudele, A.; Zaffina, S.; Denaro, M.; Smeriglio, A.; Trombetta, D. The antioxidant effects of hydroxytyrosol and vitamin e on pediatric nonalcoholic fatty liver disease, in a clinical trial: A new treatment? Antioxid. Redox Signal. 2019, 31, 127–133. [Google Scholar] [CrossRef]
- Kenzaoui, B.H.; Vilà, M.R.; Miquel, J.M.; Cengelli, F.; Juillerat-Jeanneret, L. Evaluation of uptake and transport of cationic and anionic ultrasmall iron oxide nanoparticles by human colon cells. Int. J. Nanomed. 2012, 7, 1275–1286. [Google Scholar]
- Pengnam, S.; Plianwong, S.; Patrojanasophon, P.; Radchatawedchakoon, W.; Yingyongnarongkul, B.E.; Opanasopit, P.; Charoensuksai, P. Synergistic effect of doxorubicin and siRNA-mediated silencing of Mcl-1 using cationic niosomes against 3D MCF-7 spheroids. Pharmaceutics 2021, 13, 550. [Google Scholar] [CrossRef]
- Xu, S.; Hermanson, D.J.; Banerjee, S.; Ghebreselasie, K.; Clayton, G.M.; Garavito, R.M.; Marnett, L.J. Oxicams bind in a novel mode to the cyclooxygenase active site via a two-water-mediated H-bonding network. J. Biol. Chem. 2014, 289, 6799–6808. [Google Scholar] [CrossRef]
- Kurumbail, R.G.; Stevens, A.M.; Gierse, J.K.; McDonald, J.J.; Stegeman, R.A.; Pak, J.Y.; Gildehaus, D.; Miyashiro, J.M.; Penning, T.D.; Seibert, K.; et al. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature 1996, 384, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Blobaum, A.L.; Marnett, L.J. Structural and functional basis of cyclooxygenase inhibition. J. Med. Chem. 2007, 50, 1425–1441. [Google Scholar] [CrossRef] [PubMed]
- Rouzer, C.A.; Marnett, L.J. Structural and chemical biology of the interaction of cyclooxygenase with substrates and non-steroidal anti-inflammatory drugs. Chem. Rev. 2020, 120, 7592–7641. [Google Scholar] [CrossRef]
- Akinloye, O.A.; Metibemu, D.S.; Akinloye, D.I.; Onigbinde, S.B.; Olaosebikan, I.A.; Florence, O.; Damilola, B.; Bolarinwa, O.A.; Olubunmi, O. Flavanones from Sorghum bicolor selectively inhibit COX-2: In-silico and in-vivo validation. Egypt J. Med. Hum. Genet. 2019, 20, 34–43. [Google Scholar] [CrossRef]
- Sun, H.; Chow, E.C.; Liu, S.; Du, Y.; Pang, K.S. The Caco-2 cell monolayer: Usefulness and limitations. Expert Opin. Drug Metab.Toxicol. 2008, 4, 395–411. [Google Scholar] [CrossRef]
- Kamiloglu, S.; Capanoglu, E.; Grootaert, C.; Van Camp, J. Anthocyanin absorption and metabolism by human intestinal Caco-2 cells–a review. Int. J. Mol. Sci. 2015, 16, 21555–21574. [Google Scholar] [CrossRef] [PubMed]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, Pharmacology and Health Benefits of Anthocyanins. Phytother. Res. 2016, 30, 1265–1286. [Google Scholar] [CrossRef]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Proanthocyanidins and hydrolysable tannins: Occurrence, dietary intake and pharmacological effects. Br. J. Pharmacol. 2017, 174, 1244–1262. [Google Scholar] [CrossRef]
- Smeriglio, A.; Calderaro, A.; Denaro, M.; Laganà, G.; Bellocco, E. Effects of Isolated Isoflavones Intake on Health. Curr. Med. Chem. 2019, 26, 5094–5107. [Google Scholar] [CrossRef]
- Gervasi, T.; Calderaro, A.; Barreca, D.; Tellone, E.; Trombetta, D.; Ficarra, S.; Smeriglio, A.; Mandalari, G.; Gattuso, G. Biotechnological applications and health-promoting properties of flavonols: An updated view. Int. J. Mol. Sci. 2022, 23, 1710. [Google Scholar] [CrossRef]
- Kim, H.; Lee, D.G. Naringin-generated ROS promotes mitochondria-mediated apoptosis in Candida albicans. IUBMB Life 2021, 73, 953–967. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation in human diseases. Trends Mol. Med. 2003, 9, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Ghani, M.A.; Barril, C.; Bedgood, D.R., Jr.; Prenzler, P.D. Measurement of antioxidant activity with the thiobarbituric acid reactive substances assay. Food Chem. 2017, 230, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Smeriglio, A.; Cornara, L.; Denaro, M.; Barreca, D.; Burlando, B.; Xiao, J.; Trombetta, D. Antioxidant and cytoprotective activities of an ancient Mediterranean Citrus (Citrus lumia Risso) albedo extract: Microscopic observations and polyphenol characterization. Food Chem. 2019, 279, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Diab, K.A.; Shafik, R.E.; Yasuda, S. In vitro antioxidant and antiproliferative activities of novel orange peel extract and it’s fractions on leukemia HL-60 cells. Asian Pac. J. Cancer Prev. 2015, 16, 7053–7060. [Google Scholar] [CrossRef] [PubMed]
- Bellocco, E.; Barreca, D.; Laganà, G.; Leuzzi, U.; Tellone, E.; Ficarra, S.; Kotyk, A.; Galtieri, A. Influence of L-rhamnosyl-D-glucosyl derivatives on properties and biological interaction of flavonoids. Mol. Cell Biochem. 2009, 321, 165–171. [Google Scholar] [CrossRef]
- Barreca, D.; Laganà, G.; Tellone, E.; Ficarra, S.; Leuzzi, U.; Galtieri, A.; Bellocco, E. Influences of flavonoids on erythrocyte membrane and metabolic implication through anionic exchange modulation. J. Membr. Biol. 2009, 230, 163–171. [Google Scholar] [CrossRef]
- Barreca, D.; Bellocco, E.; Caristi, C.; Leuzzi, U.; Gattuso, G. Elucidation of the flavonoid and furocoumarin composition and radical-scavenging activity of green and ripe chinotto (Citrus myrtifolia Raf.) fruit tissues, leaves and seeds. Food Chem. 2011, 129, 1504–1512. [Google Scholar] [CrossRef]
- Barreca, D.; Bisignano, C.; Ginestra, G.; Bisignano, G.; Bellocco, E.; Leuzzi, U.; Gattuso, G. Polymethoxylated, C- and O-glycosyl flavonoids in tangelo (Citrus reticulata × Citrus paradisi) juice and their influence on antioxidant properties. Food Chem. 2013, 141, 1481–1488. [Google Scholar] [CrossRef]
- Barreca, D.; Gattuso, G.; Laganà, G.; Leuzzi, U.; Bellocco, E. C- and O-glycosyl flavonoids in Sanguinello and Tarocco blood orange (Citrus sinensis (L.) Osbeck) juice: Identification and influence on antioxidant properties and acetylcholinesterase activity. Food Chem. 2016, 196, 619–627. [Google Scholar] [CrossRef]
- Lieder, B.; Hoi, J.K.; Holik, A.K.; Geissler, K.; Hans, J.; Friedl, B.; Liszt, K.; Krammer, G.E.; Ley, J.P.; Somoza, V. The flavanone homoeriodictyol increases SGLT-1-mediated glucose uptake but decreases serotonin release in differentiated Caco-2 cells. PLoS ONE 2017, 12, e0171580. [Google Scholar] [CrossRef] [PubMed]
- Gauer, J.S.; Tumova, S.; Lippiat, J.D.; Kerimi, A.; Williamson, G. Differential patterns of inhibition of the sugar transporters GLUT2, GLUT5 and GLUT7 by flavonoids. Biochem. Pharmacol. 2018, 152, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Kerimi, A.; Gauer, J.S.; Crabbe, S.; Cheah, J.W.; Lau, J.; Walsh, R.; Cancalon, P.F.; Williamson, G. Effect of the flavonoid hesperidin on glucose and fructose transport, sucrase activity and glycaemic response to orange juice in a crossover trial on healthy volunteers. Br. J. Nutr. 2019, 121, 782–792. [Google Scholar] [CrossRef]
- Zhang, H.; Hassan, Y.I.; Liu, R.; Mats, L.; Yang, C.; Liu, C.; Tsao, R. Molecular mechanisms underlying the absorption of aglycone and glycosidic flavonoids in a Caco-2 BBe1 cell model. ACS Omega 2020, 5, 10782–10793. [Google Scholar] [CrossRef] [PubMed]
- Tejada, S.; Pinya, S.; Martorell, M.; Capó, X.; Tur, J.A.; Pons, A.; Sureda, A. Potential anti-inflammatory effects of hesperidin from the genus citrus. Curr. Med. Chem. 2018, 25, 4929–4945. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Fokou, P.V.T.; Sharifi-Rad, M.; Zucca, P.; Pezzani, R.; Martins, N.; Sharifi-Rad, J. The therapeutic potential of naringenin: A review of clinical trials. Pharmaceuticals 2019, 12, 11. [Google Scholar] [CrossRef]
- Ferreira de Oliveira, J.M.P.; Santos, C.; Fernandes, E. Therapeutic potential of hesperidin and its aglycone hesperetin: Cell cycle regulation and apoptosis induction in cancer models. Phytomedicine 2020, 73, 152887. [Google Scholar] [CrossRef]
- Arafah, A.; Rehman, M.U.; Mir, T.M.; Wali, A.F.; Ali, R.; Qamar, W.; Khan, R.; Ahmad, A.; Aga, S.S.; Alqahtani, S.; et al. Multi-therapeutic potential of naringenin (4′,5,7-trihydroxyflavonone): Experimental evidence and mechanisms. Plants 2020, 9, 1784. [Google Scholar] [CrossRef]
- Smeriglio, A.; Denaro, M.; D’Angelo, V.; Germanò, M.P.; Trombetta, D. Antioxidant, anti-inflammatory and anti-angiogenic properties of Citrus lumia juice. Front. Pharmacol. 2020, 11, 593506. [Google Scholar] [CrossRef]
- Giménez-Bastida, J.A.; Martínez-Florensa, M.; Espín, J.C.; Tomás-Barberán, F.A.; García-Conesa, M.T. A Citrus extract containing flavanones represses plasminogen activator inhibitor-1 (PAI-1) expression and regulates multiple inflammatory, tissue repair, and fibrosis genes in human colon fibroblasts. J. Agric. Food Chem. 2009, 57, 9305–9315. [Google Scholar] [CrossRef]
- Kobayashi, S.; Tanabe, S.; Sugiyama, M.; Konishi, Y. Transepithelial transport of hesperetin and hesperidin in intestinal Caco-2 cell monolayers. Biochim. Biophys. Acta 2008, 1778, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Manthey, J.A.; Grohmann, K. Concentrations of hesperidin and other orange peel flavonoids in Citrus processing byproducts. J. Agric. Food Chem. 1996, 44, 811–814. [Google Scholar] [CrossRef]
- Parhiz, H.; Roohbakhsh, A.; Soltani, F.; Rezaee, R.; Iranshahi, M. Antioxidant and anti-inflammatory properties of the Citrus flavonoids hesperidin and hesperetin: An updated review of their molecular mechanisms and experimental models. Phytother. Res. 2015, 29, 323–331. [Google Scholar] [CrossRef] [PubMed]
- López-Posadas, R.; Ballester, I.; Mascaraque, C.; Suárez, M.D.; Zarzuelo, A.; Martínez-Augustin, O.; Sánchez de Medina, F. Flavonoids exert distinct modulatory actions on cyclooxygenase 2 and NF-kappaB in an intestinal epithelial cell line (IEC18). Br. J. Pharmacol. 2010, 160, 1714–1726. [Google Scholar] [CrossRef] [PubMed]






| COX-133-583 Homology Model | COX-233-583 Homology Model | Experimental COX-1 (PDB ID 6Y3C) | Experimental COX-2 (PDB ID 5IKR) | |
|---|---|---|---|---|
| Residues in most favored regions | 89.8% | 89.7% | 88.3% | 90.0% |
| Residues in additional allowed regions | 9.8% | 10.1% | 11.7% | 9.8% |
| Residues in generously allowed regions | 0.4% | 0.0% | 0.0% | 0.1% |
| Residues in disallowed regions | 0% | 0.2% | 0.0% | 0.1% |
| Sample | Inhibitory Activity (%) | |
|---|---|---|
| COX-1 | COX-2 | |
| HED | 53.41 ± 0.21 a,c | 55.18 ± 0.28 a,b |
| NHE | 50.82 ± 0.28 a,d | 55.74 ± 0.42 a,b |
| HET | 55.51 ± 0.35 a,d | 58.30 ± 0.35 a,b |
| NER | 1.21 ± 0.02 a,b,d | 78.0 ± 0.14 a,b |
| ERI | 3.12 ± 0.01 a,b,d | 56.89 ± 0.08 a,b |
| DFM | 49.80 ± 0.18 a,d | 82.45 ± 0.15 a |
| NIM | 0.30 ± 0.01 b,d | 87.93 ± 0.12 |
| Residue * | Residue Type | HED | ERI | NHE | NER | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| COX-1 | COX-2 | COX-1 | COX-2 | COX-1 | COX-2 | COX-1 | COX-2 | COX-1 | COX-2 | |
| 82 | Arg | Lys | WHB | HB | HB | |||||
| 83 | Pro | Pro | HB | WHB | HB | |||||
| 88 | Thr | Val | WHB | |||||||
| 115 | Leu | Tyr | HB | |||||||
| 116 | Val | Val | WHB | WHB | ||||||
| 119 | Val | Ser | WHB | HB | WHB | |||||
| 120 | Arg | Arg | HB, πC | WHB | HB | HB, WHB | WHB, πC | HB, πC | HB, WHB, πC | |
| 352 | Leu | Leu | WHB | |||||||
| 355 | Tyr | Tyr | ππ | HB | HB, WHB | ππ | ππ | ππ | ||
| 385 | Tyr | Tyr | WHB | |||||||
| 470 | Phe | Phe | WHB | |||||||
| 513 | His | Arg | HB | HB | ||||||
| 522 | Met | Met | HB | |||||||
| 524 | Glu | Glu | HB | WHB | WHB | HB | HB | HB | ||
| 530 | Ser | Ser | WHB | |||||||
| Ligand | ΔECOX-2-COX-1 a |
|---|---|
| HED | −2.74 |
| ERI | −38.83 |
| NHE | 3.22 |
| NER | −16.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smeriglio, A.; Iraci, N.; Denaro, M.; Mandalari, G.; Giofrè, S.V.; Trombetta, D. Synergistic Combination of Citrus Flavanones as Strong Antioxidant and COX-Inhibitor Agent. Antioxidants 2023, 12, 972. https://doi.org/10.3390/antiox12040972
Smeriglio A, Iraci N, Denaro M, Mandalari G, Giofrè SV, Trombetta D. Synergistic Combination of Citrus Flavanones as Strong Antioxidant and COX-Inhibitor Agent. Antioxidants. 2023; 12(4):972. https://doi.org/10.3390/antiox12040972
Chicago/Turabian StyleSmeriglio, Antonella, Nunzio Iraci, Marcella Denaro, Giuseppina Mandalari, Salvatore Vincenzo Giofrè, and Domenico Trombetta. 2023. "Synergistic Combination of Citrus Flavanones as Strong Antioxidant and COX-Inhibitor Agent" Antioxidants 12, no. 4: 972. https://doi.org/10.3390/antiox12040972