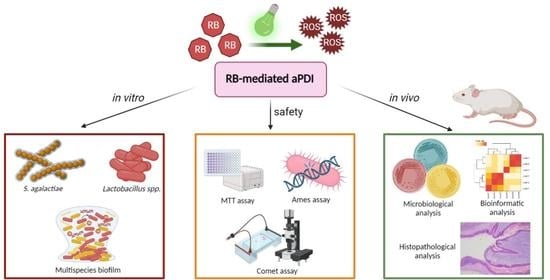
Antimicrobial Photodynamic Inactivation: An Alternative for Group B Streptococcus Vaginal Colonization in a Murine Experimental Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Media
2.2. Photosensitizing Agents
2.3. Light Sources
2.4. Photodynamic Inactivation of Planktonic Cultures
2.5. Microtiter Plate Multispecies Biofilm Culture and Photodynamic Inactivation
2.6. Photo- and Cytotoxicity Assays Based on MTT
2.7. Analysis of Real-Time Cell-Growth Dynamics
2.8. Procariotic Mutagenicity Assay–Ames Test
2.9. Eucariotic Mutagenicity Assay–Comet Test
2.10. In Vivo Experiments
2.11. Microbiological Analysis of In Vivo Experiments
2.12. Preparation of Samples for DNA Sequencing Analysis
2.13. Analysis of Sequencing Data
2.14. Histopathological Analysis
2.15. Statistical Analysis
3. Results
3.1. Streptococcus Agalactiae Can Be Eradicated with RB-Mediated aPDI in Planktonic Culture In Vitro
3.2. RB-Mediated aPDI Is Less Effective against Human Vaginal Lactobacilli Than against S. agalactiae In Vitro
3.3. RB-Mediated aPDI Inactivates S. agalactiae in Multispecies Biofilm Culture
3.4. RB-Mediated aPDI with Low Concentrations of RB Is Not Toxic for Eukaryotic Cells
3.5. RB-Mediated aPDI in the Ames Test Does Not Show Mutagenicity in Procaryotic Cells
3.6. RB-Mediated aPDI in Comet Assay Does Not Show High Mutagenicity in Eucaryotic Cells
3.7. RB-Mediated aPDI Can Inactivate S. agalactiae In Vivo in the Murine Model
3.8. The Histopathological Analysis Does Not Indicate the Negative Impact of RB-Mediated aPDI
3.9. S. agalactiae Colonization Affects the Vaginal Microbiome Composition
3.10. aPDI Impacts the Abundance of the Most Prevalent Species in Mouse Vaginas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Infectious Diseases of the Fetus and Newborn E-Book—Jack S. Remington, Christopher B. Wilson, Victor Nizet, Jerome O. Klein, Yvonne Maldonado—Google Książki. Available online: https://books.google.pl/books?hl=pl&lr=&id=5ECgVBNQDUMC&oi=fnd&pg=PP1&dq=Wilson,+C.+B.,+Nizet,+V.,+Malsonado,+Y.,+Remington,+J.+S.+%26+Klein+J.+O.+Infectious+Diseases+of+the+Fetus+and+Newborn+Infant+8th+edn+Saunders+Elsevier,+2016&ots=TJfboIZo_5&sig=BRNSI7zhywMyrr22bC1MVMNAu90&redir_esc=y#v=onepage&q&f=false (accessed on 22 February 2023).
- Prevention of Group B Streptococcal Early-Onset Disease in Newborns: ACOG Committee Opinion, Number 797. Obstet. Gynecol. 2020, 135, e51–e72. [CrossRef] [PubMed]
- Lamagni, T.L.; Keshishian, C.; Efstratiou, A.; Guy, R.; Henderson, K.L.; Broughton, K.; Sheridan, E. Emerging Trends in the Epidemiology of Invasive Group B Streptococcal Disease in England and Wales, 1991–2010. Clin. Infect. Dis. 2013, 57, 682–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okike, I.O.; Ribeiro, S.; Ramsay, M.E.; Heath, P.T.; Sharland, M.; Ladhani, S.N. Trends in bacterial, mycobacterial, and fungal meningitis in England and Wales 2004–11: An observational study. Lancet Infect. Dis. 2014, 14, 301–307. [Google Scholar] [CrossRef]
- Kohli-Lynch, M.; Russell, N.J.; Seale, A.C.; Dangor, Z.; Tann, C.J.; Baker, C.J.; Bartlett, L.; Cutland, C.; Gravett, M.G.; Heath, P.T.; et al. Neurodevelopmental Impairment in Children after Group B Streptococcal Disease Worldwide: Systematic Review and Meta-analyses. Clin. Infect. Dis. 2017, 65, S190–S199. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, Y.; Wang, Y.; Murray, C.K.; Hamblin, M.R.; Hooper, D.C.; Dai, T. Antimicrobial blue light inactivation of pathogenic microbes: State of the art. Drug Resist. Updat. 2017, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Nakonieczna, J.; Wozniak, A.; Pieranski, M.; Rapacka-Zdonczyk, A.; Ogonowska, P.; Grinholc, M. Photoinactivation of ESKAPE pathogens: Overview of novel therapeutic strategy. Future Med. Chem. 2019, 11, 443–461. [Google Scholar] [CrossRef] [PubMed]
- St. Denis, T.G.; Dai, T.; Izikson, L.; Astrakas, C.; Anderson, R.R.; Hamblin, M.R.; Tegos, G.P. All you need is light. Virulence 2011, 2, 509–520. [Google Scholar] [CrossRef]
- Kharkwal, G.B.; Sharma, S.K.; Huang, Y.Y.; Dai, T.; Hamblin, M.R. Photodynamic therapy for infections: Clinical applications. Lasers Surg. Med. 2011, 43, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Wainwright, M.; Maisch, T.; Nonell, S.; Plaetzer, K.; Almeida, A.; Tegos, G.P.; Hamblin, M.R. Photoantimicrobials—Are we afraid of the light? Lancet Infect. Dis. 2017, 17, e49–e55. [Google Scholar] [CrossRef]
- Wainwright, M. Photodynamic antimicrobial chemotherapy (PACT). J. Antimicrob. Chemother. 1998, 42, 13–28. [Google Scholar] [CrossRef]
- Baptista, M.S.; Cadet, J.; Di Mascio, P.; Ghogare, A.A.; Greer, A.; Hamblin, M.R.; Lorente, C.; Nunez, S.C.; Ribeiro, M.S.; Thomas, A.H.; et al. Type I and Type II Photosensitized Oxidation Reactions: Guidelines and Mechanistic Pathways. Photochem. Photobiol. 2017, 93, 912–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinholc, M.; Rapacka-Zdonczyk, A.; Rybak, B.; Szabados, F.; Bielawski, K.P. Multiresistant Strains Are as Susceptible to Photodynamic Inactivation as Their Naïve Counterparts: Protoporphyrin IX-Mediated Photoinactivation Reveals Differences between Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus Strains. Photomed. Laser Surg. 2014, 32, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fila, G.; Kawiak, A.; Grinholc, M.S. Blue light treatment of pseudomonas aeruginosa: Strong bactericidal activity, synergism with antibiotics and inactivation of virulence factors. Virulence 2017, 8, 938–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakonieczna, J.; Wolnikowska, K.; Ogonowska, P.; Neubauer, D.; Bernat, A.; Kamysz, W. Rose bengal-mediated photoinactivation of multidrug resistant Pseudomonas aeruginosa is enhanced in the presence of antimicrobial peptides. Front. Microbiol. 2018, 9, 1949. [Google Scholar] [CrossRef]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Quantum Yields for the Photosensitized Formation of the Lowest Electronically Excited Singlet State of Molecular Oxygen in Solution. J. Phys. Chem. Ref. Data 2009, 22, 113. [Google Scholar] [CrossRef] [Green Version]
- Kurosu, M.; Mitachi, K.; Yang, J.; Pershing, E.V.; Horowitz, B.D.; Wachter, E.A.; Lacey, J.W.; Ji, I.; Rodrigues, Y.; Antibacterial, D.J.; et al. Antibacterial Activity of Pharmaceutical-Grade Rose Bengal: An Application of a Synthetic Dye in Antibacterial Therapies. Molecules 2022, 27, 322. [Google Scholar] [CrossRef]
- Nakonechny, F.; Barel, M.; David, A.; Koretz, S.; Litvak, B.; Ragozin, E.; Etinger, A.; Livne, O.; Pinhasi, Y.; Gellerman, G.; et al. Dark Antibacterial Activity of Rose Bengal. Int. J. Mol. Sci. 2019, 20, 3196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanerio, N.; Stijnen, M.; De Mol, B.A.J.M.; Kock, L.M. Biomedical Applications of Photo- and Sono-Activated Rose Bengal: A Review. Photobiomodulation Photomed. Laser Surg. 2019, 37, 383–394. [Google Scholar] [CrossRef] [Green Version]
- Hirose, M.; Yoshida, Y.; Horii, K.; Hasegawa, Y.; Shibuya, Y. Efficacy of antimicrobial photodynamic therapy with Rose Bengal and blue light against cariogenic bacteria. Arch. Oral Biol. 2021, 122, 105024. [Google Scholar] [CrossRef]
- Pieranski, M.K.; Rychlowski, M.; Grinholc, M. Optimization of Streptococcus agalactiae Biofilm Culture in a Continuous Flow System for Photoinactivation Studies. Pathogens 2021, 10, 1212. [Google Scholar] [CrossRef]
- Michalska, K.; Rychłowski, M.; Krupińska, M.; Szewczyk, G.; Sarna, T.; Nakonieczna, J. Gallium Mesoporphyrin IX-Mediated Photodestruction: A Pharmacological Trojan Horse Strategy to Eliminate Multidrug-Resistant Staphylococcus aureus. Mol. Pharm. 2022, 19, 1434–1448. [Google Scholar] [CrossRef] [PubMed]
- Simon Andrews Babraham Bioinformatics—FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 January 2023).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Nekrutenko, A.; Grüning, B.A.; Blankenberg, D.; Goecks, J.; Schatz, M.C.; Ostrovsky, A.E.; Mahmoud, A.; Lonie, A.J.; Syme, A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Abachi, S.; Lee, S.; Vasantha Rupasinghe, H.P.; Battino, M.; Niki, E.; Quiles, J.L. Molecular Mechanisms of Inhibition of Streptococcus Species by Phytochemicals. Molecules 2016, 21, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhouioui, M.; Boulila, A.; Jemli, M.; Schiets, F.; Casabianca, H.; Zina, M.S. Fatty Acids Composition and Antibacterial Activity of Aristolochia longa L. and Bryonia dioïca Jacq. Growing Wild in Tunisia. J. Oleo Sci. 2016, 65, 655–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moncla, B.J.; Pryke, K.; Isaacs, C.E. Killing of Neisseria gonorrhoeae, Streptococcus agalactiae (group B streptococcus), Haemophilus ducreyi, and vaginal Lactobacillus by 3-O-octyl-sn-glycerol. Antimicrob. Agents Chemother. 2008, 52, 1577–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavaco, C.K.; Patras, K.A.; Zlamal, J.E.; Thoman, M.L.; Morgan, E.L.; Sanderson, S.D.; Dorana, K.S. A novel C5a-derived immunobiotic peptide reduces Streptococcus agalactiae colonization through targeted bacterial killing. Antimicrob. Agents Chemother. 2013, 57, 5492–5499. [Google Scholar] [CrossRef] [Green Version]
- Ohlsson, A.; Shah, V.S.; Stade, B.C. Vaginal chlorhexidine during labour to prevent early-onset neonatal group B streptococcal infection. Cochrane Database Syst. Rev. 2014, 12, CD003520. [Google Scholar] [CrossRef]
- Falagas, M.E.; Betsi, G.I.; Athanasiou, S. Probiotics for the treatment of women with bacterial vaginosis. Clin. Microbiol. Infect. 2007, 13, 657–664. [Google Scholar] [CrossRef] [Green Version]
- Homayouni, A.; Bastani, P.; Ziyadi, S.; Mohammad-Alizadeh-Charandabi, S.; Ghalibaf, M.; Mortazavian, A.M.; Mehrabany, E.V. Effects of probiotics on the recurrence of bacterial vaginosis: A review. J. Low. Genit. Tract Dis. 2014, 18, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Sellera, F.P.; Sabino, C.P.; Ribeiro, M.S.; Gargano, R.G.; Benites, N.R.; Melville, P.A.; Pogliani, F.C. In vitro photoinactivation of bovine mastitis related pathogens. Photodiagnosis Photodyn. Ther. 2016, 13, 276–281. [Google Scholar] [CrossRef]
- Brasel, M.; Pieranski, M.; Grinholc, M. An extended logistic model of photodynamic inactivation for various levels of irradiance using the example of Streptococcus agalactiae. Sci. Rep. 2020, 10, 14168. [Google Scholar] [CrossRef]
- Pieranski, M.; Sitkiewicz, I.; Grinholc, M. Increased photoinactivation stress tolerance of Streptococcus agalactiae upon consecutive sublethal phototreatments. Free Radic. Biol. Med. 2020, 160, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Rapacka-Zdonczyk, A.; Wozniak, A.; Pieranski, M.; Woziwodzka, A.; Bielawski, K.P.; Grinholc, M. Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinholc, M.; Rodziewicz, A.; Forys, K.; Rapacka-Zdonczyk, A.; Kawiak, A.; Domachowska, A.; Golunski, G.; Wolz, C.; Mesak, L.; Becker, K.; et al. Fine-tuning recA expression in Staphylococcus aureus for antimicrobial photoinactivation: Importance of photo-induced DNA damage in the photoinactivation mechanism. Appl. Microbiol. Biotechnol. 2015, 99, 9161–9176. [Google Scholar] [CrossRef] [Green Version]
- Sabbahi, S.; Ben Ayed, L.; Jemli, M. Staphylococcus aureus photodynamic inactivation mechanisms by rose bengal: Use of antioxidants and spectroscopic study. Appl. Water Sci. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.F.; Borges, A.; Freitas, C.F.; Hioka, N.; Mikcha, J.M.G.; Simões, M. Antimicrobial Photodynamic Inactivation Mediated by Rose Bengal and Erythrosine Is Effective in the Control of Food-Related Bacteria in Planktonic and Biofilm States. Molecules 2018, 23, 2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubey, T.; Gorantla, N.V.; Chandrashekara, K.T.; Chinnathambi, S. Photodynamic exposure of Rose-Bengal inhibits Tau aggregation and modulates cytoskeletal network in neuronal cells. Sci. Rep. 2020, 10, 12380. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, J.H.; Liu, Q.; Huang, H.; Chen, M.; Li, K.; Li, C.; Yu, X.F.; Chu, P.K. Rose-bengal-conjugated gold nanorods for in vivo photodynamic and photothermal oral cancer therapies. Biomaterials 2014, 35, 1954–1966. [Google Scholar] [CrossRef]
- Panzarini, E.; Inguscio, V.; Fimia, G.M.; Dini, L. Rose Bengal Acetate PhotoDynamic Therapy (RBAc-PDT) Induces Exposure and Release of Damage-Associated Molecular Patterns (DAMPs) in Human HeLa Cells. PLoS ONE 2014, 9, e105778. [Google Scholar] [CrossRef]
- Amescua, G.; Arboleda, A.; Nikpoor, N.; Durkee, H.; Relhan, N.; Aguilar, M.C.; Flynn, H.W.; Miller, D.; Parel, J.M. Rose Bengal Photodynamic Antimicrobial Therapy: A Novel Treatment for Resistant Fusarium Keratitis. Cornea 2017, 36, 1141. [Google Scholar] [CrossRef]
- Komagoe, K.; Kato, H.; Inoue, T.; Katsu, T. Continuous real-time monitoring of cationic porphyrin-induced photodynamic inactivation of bacterial membrane functions using electrochemical sensors. Photochem. Photobiol. Sci. 2011, 10, 1181–1188. [Google Scholar] [CrossRef]
- Alarcón, E.; Edwards, A.M.; Aspée, A.; Borsarelli, C.D.; Lissi, E.A. Photophysics and photochemistry of rose bengal bound to human serum albumin. Photochem. Photobiol. Sci. 2009, 8, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Vatansever, F.; de Melo, W.C.M.A.; Avci, P.; Vecchio, D.; Sadasivam, M.; Gupta, A.; Chandran, R.; Karimi, M.; Parizotto, N.A.; Yin, R.; et al. Antimicrobial strategies centered around reactive oxygen species—Bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 2013, 37, 955–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosini, R.; Margarit, I. Biofilm formation by Streptococcus agalactiae: Influence of environmental conditions and implicated virulence factor. Front. Cell. Infect. Microbiol. 2015, 5, 2013–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauschild, T.; Schwarz, S. Differentiation of Staphylococcus sciuri Strains Isolated from Free-Living Rodents and Insectivores. J. Vet. Med. Ser. B 2003, 50, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Argudín, M.A.; Feßler, A.T.; Hauschild, T.; Schwarz, S.; Butaye, P. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 2014, 171, 342–356. [Google Scholar] [CrossRef]
- Hagner, S.; Harb, H.; Zhao, M.; Stein, K.; Holst, O.; Ege, M.J.; Mayer, M.; Matthes, J.; Bauer, J.; Von Mutius, E.; et al. Farm-derived Gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy 2013, 68, 322–329. [Google Scholar] [CrossRef]
- Guerrero-Beltrán, C.; Garcia-Heredia, I.; Ceña-Diez, R.; Rodriguez-Izquierdo, I.; Serramía, M.J.; Martinez-Hernandez, F.; Lluesma-Gomez, M.; Martinez-Garcia, M.; Muñoz-Fernández, M.Á. Cationic Dendrimer G2-S16 Inhibits Herpes Simplex Type 2 Infection and Protects Mice Vaginal Microbiome. Pharmaceutics 2020, 12, 515. [Google Scholar] [CrossRef]
- Noguchi, K.; Tsukumi, K.; Urano, T. Qualitative and quantitative differences in normal vaginal flora of conventionally reared mice, rats, hamsters, rabbits, and dogs. Comp. Med. 2003, 53, 404–412. Available online: https://pubmed.ncbi.nlm.nih.gov/14524417/ (accessed on 24 March 2023).







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierański, M.K.; Kosiński, J.G.; Szymczak, K.; Sadowski, P.; Grinholc, M. Antimicrobial Photodynamic Inactivation: An Alternative for Group B Streptococcus Vaginal Colonization in a Murine Experimental Model. Antioxidants 2023, 12, 847. https://doi.org/10.3390/antiox12040847
Pierański MK, Kosiński JG, Szymczak K, Sadowski P, Grinholc M. Antimicrobial Photodynamic Inactivation: An Alternative for Group B Streptococcus Vaginal Colonization in a Murine Experimental Model. Antioxidants. 2023; 12(4):847. https://doi.org/10.3390/antiox12040847
Chicago/Turabian StylePierański, Michał K., Jan G. Kosiński, Klaudia Szymczak, Piotr Sadowski, and Mariusz Grinholc. 2023. "Antimicrobial Photodynamic Inactivation: An Alternative for Group B Streptococcus Vaginal Colonization in a Murine Experimental Model" Antioxidants 12, no. 4: 847. https://doi.org/10.3390/antiox12040847