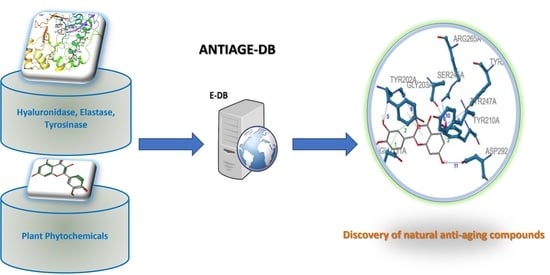
ANTIAGE-DB: A Database and Server for the Prediction of Anti-Aging Compounds Targeting Elastase, Hyaluronidase, and Tyrosinase
Abstract
:1. Introduction
2. Enzymes Related to Early Skin-Aging
2.1. Human Neutrophil Elastase (HNE)-A Serine Protease
2.1.1. Natural Sources as HNE Inhibitors
2.1.2. Natural Secondary Metabolites as HNE Inhibitors
2.2. Hyaluronidase (Hyal)—A Glycosyl Hydrolase
2.2.1. Structure of Hyals
2.2.2. Natural Sources as Hyal Inhibitors
2.2.3. Natural Secondary Metabolites as Hyaluronidase Inhibitors
2.3. Tyrosinase—A Polyphenol Oxidase
2.3.1. Structure of Tyrosinase
2.3.2. Natural Sources as Tyr Inhibitors
2.3.3. Natural Secondary Metabolites as Tyr Inhibitors
3. Software Development: The Anti-Age Database
3.1. The Anti-Age Database (ANTIAGE-DB)
3.2. Methods
3.2.1. Ligand Based Virtual Screening (LBVS)
3.2.2. Structure Based Virtual Screening (SBVS)
3.3. Anti-Age Database Design
3.3.1. Database Structure and Content
3.3.2. Submitting Calculations
3.4. Implementation and Web Access
4. Validation of ANTIAGE-DB
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Li, X. Anti-aging cosmetics and its efficacy assessment methods. IOP Conf. Ser. Mater. Sci. Eng. 2015, 87, 012043. [Google Scholar] [CrossRef] [Green Version]
- Heck, D.E.; Vetrano, A.M.; Mariano, T.M.; Laskin, J.D. UVB light stimulates production of reactive oxygen species—Unexpected role for catalase. J. Biol. Chem. 2003, 278, 22432–22436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinnerthaler, M.; Bischof, J.; Streubel, M.K.; Trost, A.; Richter, K. Oxidative stress in aging human skin. Biomolecules 2015, 5, 545–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Necas, J.; Bartosikova, L.; Brauner, P.; Kolar, J. Hyaluronic acid (hyaluronan): A review. Vet. Med. 2008, 53, 397–411. [Google Scholar] [CrossRef] [Green Version]
- Chang, T.S. An updated review of tyrosinase inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef] [Green Version]
- Mainka, M.; Czerwińska, M.E.; Osińska, E.; Ziaja, M.; Bazylko, A. Screening of antioxidative properties and inhibition of inflammation-linked enzymes by aqueous and ethanolic extracts of plants traditionally used in wound healing in Poland. Antioxidants 2021, 10, 698. [Google Scholar] [CrossRef]
- Kim, K.; Lee, Y.S.; Kim, N.; Choi, H.D.; Lim, K.M. 5G Electromagnetic Radiation Attenuates Skin Melanogenesis In Vitro by Suppressing ROS Generation. Antioxidants 2022, 11, 1449. [Google Scholar] [CrossRef]
- Myung, C.H.; Kim, K.; Park, J.I.; Lee, J.E.; Lee, J.A.; Hong, S.C.; Lim, K.-M.; Hwang, J.S. 16-Kauren-2-beta-18, 19-triol inhibits melanosome transport in melanocytes by down-regulation of melanophilin expression. J. Dermatol. Sci. 2020, 97, 101–108. [Google Scholar] [CrossRef]
- Southam, C.M. Effects of extracts of western red-cedar heartwood on certain wood-decaying fungi in culture. Phytophathology 1943, 33, 517–524. [Google Scholar]
- Calabrese, E.J.; Dhawan, G.; Kapoor, R.; Iavicoli, I.; Calabrese, V. What is hormesis and its relevance to healthy aging and longevity? Biogerontology 2015, 16, 693–707. [Google Scholar] [CrossRef]
- Calabrese, E.J. Hormesis: A fundamental concept in biology. Microb. Cell 2014, 1, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Rattan, S.I.S. Aging, anti-aging, and hormesis. Mech. Ageing Dev. 2004, 125, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Minois, N. Applying hormesis in aging research and therapy: A commentary. Hum. Exp. Toxicol. 2001, 20, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Heiflick, L. The future of ageing. Nature 2000, 408, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. Hormetic Mechanisms. Crit. Rev. Toxicol. 2013, 43, 580–606. [Google Scholar] [CrossRef]
- Bieth, J.G. The elastases. J. Soc. Biol. 2001, 195, 173–179. [Google Scholar] [CrossRef]
- Takahashi, H.; Nukiwa, T.; Yoshimura, K.; Quick, C.D.; States, D.J.; Holmes, M.D.; Whang-Peng, J.; Knutsen, T.; Crystal, R.G. Structure of the human neutrophil elastase gene. J. Biol. Chem. 1988, 263, 14739–14747. [Google Scholar] [CrossRef]
- Melzig, M.F.; Löser, B.; Ciesielski, S. Inhibition of neutrophil elastase activity by phenolic compounds from plants. Pharmazie 2001, 56, 967–970. [Google Scholar]
- Siedle, B.; Cisielski, S.; Murillo, R.; Lo, B.; Castro, V.; Klaas, C.A.; Hucke, O.; Labahn, A.; Melzig, M.F.; Merfort, I.; et al. Sesquiterpene Lactones as Inhibitors of Human Neutrophil Elastase. Bioorg. Med. Chem. 2002, 10, 2855–2861. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Uyama, H.; Kobayashi, S. Inhibition effects of (+)-catechin-aldehyde polycondensates on proteinases causing proteolytic degradation of extracellular matrix. Biochem. Biophys. Res. Commun. 2004, 320, 256–261. [Google Scholar] [CrossRef]
- Thring, T.S.; Hili, P.; Naughton, D.P. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement. Altern. Med. 2009, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Belaaouaj, A.; Kim, K.S.; Shapiro, S.D. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 2000, 289, 1185–1188. [Google Scholar] [CrossRef]
- Ying, Q.L.; Rinehart, A.R.; Simon, S.R.; Cheronis, J.C. Inhibition of human leucocyte elastase by ursolic acid. Evidence for a binding site for pentacyclic triterpenes. Biochem. J. 1991, 277, 521–526. [Google Scholar] [CrossRef] [Green Version]
- Bode, W.; Edgar Meyer, J.C.P., Jr. Perspectives in Biochemistry. Human Leukocyte and PorcinePancreatic Elastase: X-ray Crystal Structures, Mechanism, Substrate Specificity. Biochemistry 1989, 28, 1951–1963. [Google Scholar] [CrossRef]
- Feng, L.; Liu, X.; Zhu, W.; Guo, F.; Wu, Y.; Wang, R.; Chen, K.; Huang, C.; Li, Y. Inhibition of human neutrophil elastase by pentacyclic triterpenes. PLoS ONE 2013, 8, e82794. [Google Scholar] [CrossRef] [Green Version]
- Tamada, T.; Kinoshita, T.; Kurihara, K.; Adachi, M.; Ohhara, T.; Imai, K.; Kuroki, R.; Tada, T. Combined high-resolution neutron and X-ray analysis of inhibited elastase confirms the active-site oxyanion hole but rules against a low-barrier hydrogen bond. J. Am. Chem. Soc. 2009, 131, 11033–11040. [Google Scholar] [CrossRef]
- Hess, G.P.; McConn, J.; Ku, E.; McConkey, G. Studies of the activity of chymotrypsin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1970, 257, 89–104. [Google Scholar]
- Wittenauer, J.; Mäckle, S.; Sußmann, D.; Schweiggert-Weisz, U.; Carle, R. Inhibitory effects of polyphenols from grape pomace extract on collagenase and elastase activity. Fitoterapia 2015, 101, 179–187. [Google Scholar] [CrossRef]
- Alasbahi, R.; Matthias, M. The in vitro inhibition of human neutrophil elastase activity by some Yemeni medicinal plants. Planta Med. 2008, 74, 981. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.K.; Kim, J.H.; Cho, J.J.; Choi, J.D. Inhibitory Effects of 150 Plant Extracts on Elastase Activity, and Their Anti-inflammatory Effects. Int. J. Cosmet. Sci. 1999, 21, 71–82. [Google Scholar] [CrossRef]
- Pinelo, M.; Rubilar, M.; Jerez, M.; Sineiro, J.; Núñez, M.J. Effect of solvent, temperature, and solvent-to-solid ratio on the total phenolic content and antiradical activity of extracts from different components of grape pomace. J. Agric. Food Chem. 2005, 53, 2111–2117. [Google Scholar] [CrossRef]
- Chiang, H.-M.; Lin, T.-J.; Chiu, C.-Y.; Chang, C.-W.; Hsu, K.-C.; Fan, P.-C.; Wen, K.-C. Coffea arabica extract and its constituents prevent photoaging by suppressing MMPs expression and MAP kinase pathway. Food Chem. Toxicol. 2011, 49, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, D.; Claus, A.; Carle, R.; Schieber, A. Polyphenol Screening of Pomace from Red and White Grape Varieties (Vitis vinifera L.) by HPLC-DAD-MS/MS. J. Agric. Food Chem. 2004, 52, 4360–4367. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Angelis, A.; Aligiannis, N.; Rosalia, M.; Abedini, A.; Bakiri, A.; Reynaud, R.; Nuzillard, J.M.; Gangloff, S.C.; Skaltsounis, A.L.; et al. In vitro dermo-cosmetic evaluation of bark extracts from common temperate trees. Planta Med. 2016, 82, 1351–1358. [Google Scholar] [CrossRef] [Green Version]
- Sumantran, V.N.; Kulkarni, A.A.; Harsulkar, A.; Wele, A.; Koppikar, S.J.; Chandwaskar, R.; Gaire, V.; Dalvi, M.; Wagh, U.V. Hyaluronidase and collagenase inhibitory activities of the herbal formulation Triphala guggulu. J. Biosci. 2007, 32, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Li, S.M.; Shen, Y.H.; Zhang, W.D. Phytochemical and biological studies of Abies species. Chem. Biodivers. 2008, 5, 56–81. [Google Scholar] [CrossRef]
- Kostova, I.; Iossifova, T. Chemical components of Fraxinus species. Fitoterapia 2007, 78, 85–106. [Google Scholar] [CrossRef]
- Nahrstedt, A.; Schmidt, M.; Jäggi, R.; Metz, J.; Khayyal, M.T. Willow bark extract: The contribution of polyphenols to the overall effect. Wien. Med. Wochenschr. 2007, 157, 348–351. [Google Scholar] [CrossRef]
- Tyagi, S.C.; Simon, S.R. Inhibitors directed to binding domains in neutrophil elastase. Biochemistry 1990, 29, 9970–9977. [Google Scholar] [CrossRef]
- Siedle, B.; Hrenn, A.; Merfort, I. Natural compounds as inhibitors of human neutrophil elastase. Planta Med. 2007, 73, 401–420. [Google Scholar] [CrossRef] [Green Version]
- Löser, B.; Kruse, S.O.; Melzig, M.F.; Nahrstedt, A. Inhibition of neutrophil elastase activity by cinnamic acid derivatives from Cimicifuga racemosa. Planta Med. 2000, 66, 751–753. [Google Scholar] [CrossRef]
- Melzig, M.F.; Löser, B.; Lobitz, G.O.; Tamayo-Castillo, G.; Merfort, I. Inhibition of granulocyte elastase activity by caffeic acid derivatives. Pharmazie 1999, 54, 712. [Google Scholar]
- Hamburger, M.; Riese, U.; Graf, H.; Melzig, M.F.; Ciesielski, S.; Baumann, D.; Dittmann, K.; Wegner, C. Constituents in Evening Primrose Oil with Radical Scavenging, Cyclooxygenase, and Neutrophil Elastase Inhibitory Activities. J. Agric. Food Chem. 2002, 50, 5533–5538. [Google Scholar] [CrossRef]
- Xing, X.; Yang, X.; Cao, Y. Study of Ellagic Acid as a Natural Elastase Inhibitor by Spectroscopic Methods. J. Appl. Spectrosc. 2016, 83, 149–155. [Google Scholar] [CrossRef]
- Rennert, B.; Melzig, M.F. Free fatty acids inhibit the activity of Clostridium histolyticum collagenase and human neutrophil elastase. Planta Med. 2002, 68, 767–769. [Google Scholar] [CrossRef]
- Bizot-Foulon Godeau, G.; Guessous, F.; Lati, E.; Rousset, G.; Roch-Arveillier, M.; Hornebeck, W.V. Inhibition of human neutrophil elastase by wheat ceramides. Int. J. Cosmet. Sci. 1995, 17, 255–264. [Google Scholar] [CrossRef]
- Sim, G.S.; Lee, B.C.; Cho, H.S.; Lee, J.W.; Kim, J.H.; Lee, D.H.; Kim, J.-H.; Pyo, H.B.; Moon, D.C.; Oh, K.W.; et al. Structure activity relationship of antioxidative property of flavonoids and inhibitory effect on matrix metalloproteinase activity in UVA-irradiated human dermal fibroblast. Arch. Pharmacol. Res. 2007, 30, 290–298. [Google Scholar] [CrossRef]
- Krenn, L.; Wollenweber, E.; Steyrleuthner, K.; Görick, C.; Melzig, M.F. Contribution of methylated exudate flavonoids to the anti-inflammatory activity of Grindelia robusta. Fitoterapia 2009, 80, 267–269. [Google Scholar] [CrossRef]
- Tan, X.; Song, Y.H.; Park, C.; Lee, K.W.; Kim, J.Y.; Kim, D.W.; Kim, K.D.; Lee, K.W.; Curtis-Long, M.J.; Park, K.H. Highly potent tyrosinase inhibitor, neorauflavane from Campylotropis hirtella and inhibitory mechanism with molecular docking. Bioorgan. Med. Chem. 2016, 24, 153–159. [Google Scholar] [CrossRef]
- Kim, J.Y.; Wang, Y.; Uddin, Z.; Song, Y.H.; Li, Z.P.; Jenis, J.; Park, K.H. Competitive neutrophil elastase inhibitory isoflavones from the roots of Flemingia philippinensis. Bioorg. Chem. 2018, 78, 249–257. [Google Scholar] [CrossRef]
- Sivamani, P.; Singaravelu, G.; Thiagarajan, V.; Jayalakshmi, T.; Kumar, G.R. Comparative molecular docking analysis of essential oil constituents as elastase inhibitors. Bioinformation 2012, 8, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Mattson, M.P.; Calabrese, V. Resveratrol commonly displays hormesis: Occurrence and biomedical significance. Hum. Exp. Toxicol. 2010, 29, 980–1015. [Google Scholar] [CrossRef] [PubMed]
- Grubauer, G.; Elias, P.M.; Feingold, K.R. Transepidermal water loss: The signal for recovery of barrier structure and function. J. Lipid Res. 1989, 30, 323–333. [Google Scholar] [CrossRef]
- Nakajima, K.; Powers, J.C.; Ashe, B.M.; Zimmerman, M. Mapping the extended substrate binding site of cathepsin G and human leukocyte elastase. Studies with peptide substrates related to the alpha 1-protease inhibitor reactive site. J. Biol. Chem. 1979, 254, 4027–4032. [Google Scholar] [CrossRef]
- Baici, A.; Diczházi, C.; Neszmélyi, A.; Móczár, E.; Hornebeck, W. Inhibition of the human leukocyte endopeptidases elastase and cathepsin G and of porcine pancreatic elastase by N-oleoyl derivatives of heparin. Biochem. Pharmacol. 1993, 46, 1545–1549. [Google Scholar] [CrossRef]
- Fujino, Y.; Ohnishi, M. Constituents of ceramide and ceramide monohexoside in rice bran. Chem. Phys. Lipids 1976, 17, 275–289. [Google Scholar] [CrossRef]
- Fujino, Y.; Ohnishi, M. Sphingolipids in wheat grain. J. Cereal Sci. 1983, 1, 159–168. [Google Scholar] [CrossRef]
- Feisst, C.; Franke, L.; Appendino, G.; Werz, O. Identification of molecular targets of the oligomeric nonprenylated acylphloroglucinols from Myrtus communis and their implication as anti-inflammatory compounds. J. Pharmacol. Exp. Ther. 2005, 315, 389–396. [Google Scholar] [CrossRef] [Green Version]
- Feisst, C.; Werz, O. Suppression of receptor-mediated Ca2+ mobilization and functional leukocyte responses by hyperforin. Biochem. Pharmacol. 2004, 67, 1531–1539. [Google Scholar] [CrossRef]
- Załuski, D.; Cieśla, Ł.; Janeczko, Z. Chapter 7—The Structure—Activity Relationships of Plant Secondary Metabolites with Antimicrobial, Free Radical Scavenging and Inhibitory Activity toward Selected Enzymes. Stud. Nat. Prod. Chem. 2015, 45, 217–249. [Google Scholar]
- Marković-Housley, Z.; Miglierini, G.; Soldatova, L.; Rizkallah, P.J.; Müller, U.; Schirmer, T. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure 2000, 8, 1025–1035. [Google Scholar] [CrossRef]
- Laurent, T.C.; Fraser, J.R. Hyaluronan. FASEB J. 1992, 6, 2397–2404. [Google Scholar] [CrossRef]
- Meyer, K. Hyaluronidases. In The Enzymes; Academic Press: New York, NY, USA, 1971; pp. 307–320. [Google Scholar]
- Stern, R. Devising a pathway for hyaluronan catabolism: Are we there yet? Glycobiology 2003, 13, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Orlando, Z.; Lengers, I.; Melzig, M.F.; Buschauer, A.; Hensel, A.; Jose, J. Autodisplay of human hyaluronidase Hyal-1 on Escherichia coli and identification of plant-derived enzyme inhibitors. Molecules 2015, 20, 15449–15468. [Google Scholar] [CrossRef] [Green Version]
- Jedrzejas, M.J.; Stern, R. Structures of vertebrate hyaluronidases and their unique enzymatic mechanism of hydrolysis. Proteins 2005, 61, 227–238. [Google Scholar] [CrossRef]
- Csoka, A.B.; Frost, G.I.; Stern, R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001, 20, 499–508. [Google Scholar] [CrossRef]
- Chao, K.L.; Muthukumar, L.; Herzberg, O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry 2007, 46, 6911–6920. [Google Scholar] [CrossRef] [Green Version]
- Lokeshwar, V.B.; Rubinowicz, D.; Schroeder, G.L.; Forgacs, E.; Minna, J.D.; Block, N.L.; Nadji, M.; Lokeshwar, B.L. Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J. Biol. Chem. 2001, 276, 11922–11932. [Google Scholar] [CrossRef] [Green Version]
- Lokeshwar, V.B.; Obek, C.; Pham, H.T.; Wei, D.; Young, M.J.; Duncan, R.C.; Soloway, M.S.; Block, N.L. Urinary hyaluronic acid and hyaluronidase: Markers for bladder cancer detection and evaluation of grade. J. Urol. 2000, 163, 348–356. [Google Scholar] [CrossRef]
- Tan, J.X.; Wang, X.Y.; Li, H.Y.; Su, X.L.; Wang, L.; Ran, L.; Zheng, K.; Ren, G.S. HYAL1 overexpression is correlated with the malignant behavior of human breast cancer. Int. J. Cancer 2011, 128, 1303–1315. [Google Scholar] [CrossRef]
- Mio, K.; Stern, R. Inhibitors of the hyaluronidases. Matrix Biol. 2002, 21, 31–37. [Google Scholar] [CrossRef]
- Jentsch, H.; Pomowski, R.; Kundt, G.; Göcke, R. Treatment of gingivitis with hyaluronan. J. Clin. Periodontol. 2003, 30, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Stern, R.; Jedrzejas, M.J. Hyaluronidases: Their genomics, structures, and mechanisms of action. Chem. Rev. 2006, 106, 818–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunnicutt, G.R.; Primakoff, P.; Myles, D.G. Sperm surface protein PH-20 is bifunctional: One activity is a hyaluronidase and a second, distinct activity is required in secondary sperm-zona binding. Biol. Reprod. 1996, 55, 80–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fayad, S.; Nehmé, R.; Tannoury, M.; Lesellier, E.; Pichon, C.; Morin, P. Macroalga Padina pavonica water extracts obtained by pressurized liquid extraction and microwave-assisted extraction inhibit hyaluronidase activity as shown by capillary electrophoresis. J. Chromatogr. A 2017, 1497, 19–27. [Google Scholar] [CrossRef]
- Bralley, E.; Greenspan, P.; Hargrove, J.L.; Hartle, D.K. Inhibition of hyaluronidase activity by select sorghum brans. J. Med. Food 2008, 11, 307–312. [Google Scholar] [CrossRef]
- Pessini, A.C.; Takao, T.T.; Cavalheiro, E.C.; Vichnewski, W.; Sampaio, S.V.; Giglio, J.R.; Arantes, E.C. A hyaluronidase from Tityus serrulatus scorpion venom: Isolation, characterization and inhibition by flavonoids. Toxicon 2001, 39, 1495–1504. [Google Scholar] [CrossRef]
- Pujiarti, R.; Ohtani, Y.; Ichura, H. Antioxidant, Anti-Hyaluronidase and Antifungal Activities of Melaleuca leucadendron Linn. Leaf Oils. J. Wood Sci. 2012, 58, 429–436. [Google Scholar]
- Załuski, D.; Olech, M.; Kuźniewski, R.; Verpoorte, R.; Nowak, R.; Smolarz, H.D. LC-ESI-MS/MS profiling of phenolics from Eleutherococcus spp. inflorescences, structure-activity relationship as antioxidants, inhibitors of hyaluronidase and acetylcholinesterase. Saudi Pharm. J. 2017, 25, 734–743. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.H.; Lee, J.H.; Kim, K.T.; Park, Y.S.; Nah, S.Y.; Ahn, D.U.; Paik, H.D. Antimicrobial Effect of 7-O-Butylnaringenin, a Novel Flavonoid, and Various Natural Flavonoids against Helicobacter Pylori Strains. Food Sci. Biotechnol. 2009, 18, 267–270. [Google Scholar] [CrossRef]
- Hertel, W.; Peschel, G.; Ozegowski, J.-H.; Müller, P.-J. Inhibitory Effects of Triterpenes and Flavonoids on the Enzymatic Activity of Hyaluronic Acid-Splitting Enzymes. Arch. Pharm. 2006, 339, 313–318. [Google Scholar] [CrossRef]
- Hisao, K.; Matsumoto, H.; Satoh, T. Inhibitory Effects of Some Natural Products on the Activation of Hyaluronidase and Their Antiallergic Actions. Chem. Pharm. Bull. 1992, 40, 1439–1442. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.J.; Yang, R.; You, J.; Qu, L.B.; Sun, Y.J. Spectroscopic and Docking Studies on the Binding of Liquiritigenin with Hyaluronidase for Antiallergic Mechanism. Scientifica 2016, 2016, 9178097. [Google Scholar] [CrossRef] [Green Version]
- Barla, F.; Higashijima, H.; Funai, S.; Sugimoto, K.; Harada, N.; Yamaji, R.; Fujita, T.; Nakano, Y.; Inui, H. Inhibitive effects of alkyl gallates on hyaluronidase and collagenase. Biosci. Biotechnol. Biochem. 2009, 73, 2335–2337. [Google Scholar] [CrossRef]
- Lee, S.Y.; Baek, N.; Nam, T.G. Natural, semisynthetic and synthetic tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 1–13. [Google Scholar] [CrossRef]
- Jackman, M.P.; Hajnal, A.; Lerch, K. Albino mutants of Streptomyces glaucescens tyrosinase. Biochem. J. 1991, 274 Pt 3, 707–713. [Google Scholar] [CrossRef]
- van Gelder, C.W.; Flurkey, W.H.; Wichers, H.J. Sequence and structural features of plant and fungal tyrosinases. Phytochemistry 1997, 45, 1309–1323. [Google Scholar] [CrossRef]
- Himmelwright, R.S.; Eickman, N.C.; Lu Bien, C.D.; Lerch, K.; Solomon, E. Chemical and Spectroscopic Studies of the Binuclear Copper Active Site of Neurospora Tyrosinase: Comparison to Hemocyanins. J. Am. Chem. Soc. 1980, 102, 7339–7344. [Google Scholar] [CrossRef]
- Magnus, K.A.; Hazes, B.; Ton-That, H.; Bonaventura, C.; Bonaventura, J.; Hol, W.G.J. Crystallographic Analysis of Oxygenated and Deoxygenated States of Arthropod Hemocyanin Shows Unusual Differences. Genet. Proteins Struct. Funct. 1994, 19, 302–309. [Google Scholar] [CrossRef]
- Volbeda, A.; Hol, W.G. Crystal Structure of Hexameric Haemocyanin from Panulirus Interruptus Refined at 3.2 A Resolution. J. Mol. Biol. 1989, 209, 249–279. [Google Scholar] [CrossRef]
- Wang, C.; Yan, S.; Huang, R.; Feng, S.; Fu, B.; Weng, X.; Zhou, X. A turn-on fluorescent probe for detection of tyrosinase activity. Analyst 2013, 138, 2825–2828. [Google Scholar] [CrossRef] [PubMed]
- Kubo, I.; Kinst-Hori, I.; Chaudhuri, S.K.; Kubo, Y.; Sánchez, Y.; Ogura, T. Flavonols from Heterotheca inuloides: Tyrosinase inhibitory activity and structural criteria. Bioorg. Med. Chem. 2000, 8, 1749–1755. [Google Scholar] [CrossRef]
- Kubo, I.; Kinst-Hori, I.; Yokokawa, Y. Tyrosinase inhibitors from Anacardium occidentale fruits. J. Nat. Prod. 1994, 57, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-X.; Kubo, I. Kinetics of Mushroom Tyrosinase Inhibition by Quercetin. J. Agric. Food Chem. 2002, 50, 4108–4112. [Google Scholar] [CrossRef] [PubMed]
- Kubo, I.; Kinst-Hori, I.; Kubo, Y.; Yamagiwa, Y.; Kamikawa, T.; Haraguchi, H. Molecular design of antibrowning agents. J. Agric. Food Chem. 2000, 48, 1393–1399. [Google Scholar] [CrossRef]
- Kubo, I.; Yokokawa, Y.; Kinst-Hori, I. Tyrosinase inhibitors from Bolivian medicinal plants. J. Nat. Prod. 1995, 58, 739–743. [Google Scholar] [CrossRef]
- Badria, F.A.; elGayyar, M.A. A new type of tyrosinase inhibitors from natural products as potential treatments for hyperpigmentation. Boll. Chim. Farm. 2001, 140, 267–271. [Google Scholar]
- Xie, L.-P.; Chen, Q.-X.; Huang, H.; Wang, H.-Z.; Zhang, R.-Q. Inhibitory effects of some flavonoids on the activity of mushroom tyrosinase. Biochemistry. 2003, 68, 487–491. [Google Scholar]
- Kubo, I.; Chen, Q.X.; Kenichi, N. Molecular Design of Antibrowning Agents: Antioxidative Tyrosinase Inhibitors. Food Chem. 2003, 81, 241–247. [Google Scholar] [CrossRef]
- No, J.K.; Soung, D.Y.; Kim, Y.J.; Shim, K.H.; Jun, Y.S.; Rhee, S.H.; Yokozawa, T.; Chung, H.Y. Inhibition of tyrosinase by green tea components. Life Sci. 1999, 65, PL241–PL246. [Google Scholar] [CrossRef]
- Kim, J.H.; Sapers, G.M.; Choi, S.W. Identification of Tyrosinase Inhibitor from Galla Rhois. Food Sci. Biotechnol. 1998, 7, 56–59. [Google Scholar]
- Parvez, S.; Kang, M.; Chung, H.-S.; Bae, H. Naturally occurring tyrosinase inhibitors: Mechanism and applications in skin health, cosmetics and agriculture industries. Phytother. Res. 2007, 21, 805–816. [Google Scholar] [CrossRef]
- Menon, S.; Fleck, R.W.; Yong, G.; Strothkamp, K.G. Benzoic acid inhibition of the alpha, beta, and gamma isozymes of Agaricus bisporus tyrosinase. Arch. Biochem. Biophys. 1990, 280, 27–32. [Google Scholar] [CrossRef]
- Kermasha, S.; Goetghebeur, M.; Monfette, A.; Metche, M.; Rovel, B. Inhibitory effects of cysteine and aromatic acids on tyrosinase activity. Phytochemistry 1993, 34, 349–353. [Google Scholar] [CrossRef]
- Jiménez, M.; Chazarra, S.; Escribano, J.; Cabanes, J.; García-Carmona, F. Competitive Inhibition of Mushroom Tyrosinase by 4-Substituted Benzaldehydes. J. Agric. Food Chem. 2001, 49, 4060–4063. [Google Scholar] [CrossRef]
- Wilcox, D.E.; Porras, A.G.; Hwang, Y.T.; Lerch, K.; Winkler, M.E.; Solomon, E.I. Substrate analog binding to the coupled binuclear copper active site in tyrosinase. J. Am. Chem. Soc. 1985, 107, 4015–4027. [Google Scholar] [CrossRef]
- Dilberger, B.; Weppler, S.; Eckert, G.P. Phenolic acid metabolites of polyphenols act as inductors for hormesis in C. elegans. Mech. Ageing Dev. 2021, 198, 111518. [Google Scholar] [CrossRef]
- Kim, Y.J. Rhamnetin attenuates melanogenesis by suppressing oxidative stress and pro-inflammatory mediators. Biol. Pharm. Bull. 2013, 36, 1341–1347. [Google Scholar] [CrossRef] [Green Version]
- Stratford, M.R.L.; Ramsden, C.A.; Riley, P.A. Mechanistic studies of the inactivation of tyrosinase by resorcinol. Bioorg. Med. Chem. 2013, 21, 1166–1173. [Google Scholar] [CrossRef]
- Tang, H.; Cui, F.; Li, H.; Huang, Q.; Li, Y. Understanding the inhibitory mechanism of tea polyphenols against tyrosinase using fluorescence spectroscopy, cyclic voltammetry, oximetry, and molecular simulations. RSC Adv. 2018, 8, 8310–8318. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.R.; Kang, S.; Deng, L.; Xiang, L.P.; Zheng, X.Q. Inhibitory effects of (-)-epigallocatechin-3-gallate on melanogenesis in ultraviolet A-induced B16 murine melanoma cell. Trop. J. Pharm. Res. 2014, 13, 1825–1831. [Google Scholar] [CrossRef]
- Jin, Y.J.; Lin, C.C.; Lu, T.M.; Li, J.H.; Chen, I.S.; Kuo, Y.H.; Ko, H.H. Chemical constituents derived from Artocarpus xanthocarpus as inhibitors of melanin biosynthesis. Phytochemistry 2015, 117, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Arung, E.T.; Shimizu, K.; Kondo, R. Structure-activity relationship of prenyl-substituted polyphenols from Artocarpus heterophyllus as inhibitors of melanin biosynthesis in cultured melanoma cells. Chem. Biodivers. 2007, 4, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.P.; Hsu, F.L.; Chen, C.S.; Chern, J.W.; Lee, M.H. Constituents from the Formosan apple reduce tyrosinase activity in human epidermal melanocytes. Phytochemistry 2007, 68, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Briganti, S.; Camera, E.; Picardo, M. Chemical and instrumental approaches to treat hyperpigmentation. Pigment Cell Res. 2003, 16, 101–110. [Google Scholar] [CrossRef]
- Khatib, S.; Nerya, O.; Musa, R.; Shmuel, M.; Tamir, S.; Vaya, J. Chalcones as potent tyrosinase inhibitors: The importance of a 2,4-substituted resorcinol moiety. Bioorg. Med. Chem. 2005, 13, 433–441. [Google Scholar] [CrossRef]
- Li, N.; Xue, M.-H.; Yao, H.; Zhu, J.-J. Reagentless biosensor for phenolic compounds based on tyrosinase entrapped within gelatine film. Anal. Bioanal. Chem. 2005, 383, 1127–1132. [Google Scholar] [CrossRef]
- Matoba, Y.; Kumagai, T.; Yamamoto, A.; Yoshitsu, H.; Sugiyama, M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J. Biol. Chem. 2006, 281, 8981–8990. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Guo, X.H.; Wang, S.S.; Li, Y.Q.; Li, G.Z.; Zhao, W.J. Screening and identification of natural ligands of tyrosinase from: Pueraria lobata Ohwi by a combination of ultrafiltration and LC-MS. Anal. Methods 2017, 9, 4858–4862. [Google Scholar] [CrossRef]
- Placines, C.; Castañeda-Loaiza, V.; Rodrigues, M.J.; Pereira, C.G.; Stefanucci, A.; Mollica, A.; Zengin, G.; Llorent-Martínez, E.J.; Castilho, P.C.; Custódio, L. Phenolic profile, toxicity, enzyme inhibition, in silico studies, and antioxidant properties of Cakile maritima scop. (brassicaceae) from Southern Portugal. Plants 2020, 9, 142. [Google Scholar] [CrossRef] [Green Version]
- Amer, M.; Metwalli, M. Topical hydroquinone in the treatment of some hyperpigmentary disorders. Int. J. Dermatol. 1998, 37, 449–450. [Google Scholar] [CrossRef]
- Draelos, Z.D. Skin lightening preparations and the hydroquinone controversy. Dermatol. Ther. 2007, 20, 308–313. [Google Scholar] [CrossRef]
- Parejo, I.; Viladomat, F.; Bastida, J.; Codina, C. A single extraction step in the quantitative analysis of arbutin in bearberry (Arctostaphylos uva-ursi) leaves by high-performance liquid chromatography. Phytochem. Anal. 2001, 12, 336–339. [Google Scholar] [CrossRef]
- Jin, Y.H.; Lee, S.J.; Chung, M.H.; Park, J.H.; Park, Y.I.; Cho, T.H.; Lee, S.K. Aloesin and arbutin inhibit tyrosinase activity in a synergistic manner via a different action mechanism. Arch. Pharm. Res. 1999, 22, 232–236. [Google Scholar] [CrossRef]
- Jones, K.; Hughes, J.; Hong, M.; Jia, Q.; Orndorff, S. Modulation of melanogenesis by aloesin: A competitive inhibitor of tyrosinase. Pigment Cell Res. 2002, 15, 335–340. [Google Scholar] [CrossRef]
- Wolverton, S.E.; Wu, J.J. Cosmetic therapy. In Comprehensive Dermatologic Drug Therapy, 2nd ed.; Wolverton, S.E., Ed.; Saunders Elsevier: Philadelphia, PA, USA, 2019; pp. 761–774. [Google Scholar]
- Batovska, D.I.; Todorova, I.T. Trends in utilization of the pharmacological potential of chalcones. Curr. Clin. Pharmacol. 2010, 5, 1–29. [Google Scholar] [CrossRef]
- Kim, S.J.; Son, K.H.; Chang, H.W.; Kang, S.S.; Kim, H.P. Tyrosinase inhibitory prenylated flavonoids from Sophora flavescens. Biol. Pharm. Bull. 2003, 26, 1348–1350. [Google Scholar] [CrossRef] [Green Version]
- Hyun, S.K.; Lee, W.-H.; Jeong, D.M.; Kim, Y.; Choi, J.S. Inhibitory effects of kurarinol, kuraridinol, and trifolirhizin from Sophora flavescens on tyrosinase and melanin synthesis. Biol. Pharm. Bull. 2008, 31, 154–158. [Google Scholar] [CrossRef] [Green Version]
- Nerya, O.; Musa, R.; Khatib, S.; Tamir, S.; Vaya, J. Chalcones as potent tyrosinase inhibitors: The effect of hydroxyl positions and numbers. Phytochemistry 2004, 65, 1389–1395. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, M.H.K.; Nguyen, H.X.; Bui, N.K.N.; Nguyen, M.T.T. Tyrosinase inhibitors from the wood of Artocarpus heterophyllus. J. Nat. Prod. 2012, 75, 1951–1955. [Google Scholar] [CrossRef]
- Wang, Y.; Curtis-Long, M.J.; Lee, B.W.; Yuk, H.J.; Kim, D.W.; Tan, X.F.; Park, K.H. Inhibition of tyrosinase activity by polyphenol compounds from Flemingia philippinensis roots. Bioorgan. Med. Chem. 2014, 22, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Bhardwaj, A.; Aggarwal, R.S.; Seeram, N.P.; Shishodia, S.; Takada, Y. Role of resveratrol in prevention and therapy of cancer: Preclinical and clinical studies. Anticancer Res. 2004, 24, 2783–2840. [Google Scholar] [PubMed]
- Shin, N.-H.; Ryu, S.Y.; Choi, E.J.; Kang, S.-H.; Chang, I.-M.; Min, K.R.; Kim, Y. Oxyresveratrol as the Potent Inhibitor on Dopa Oxidase Activity of Mushroom Tyrosinase. Biochem. Biophys. Res. Commun. 1998, 243, 801–803. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Kondo, R.; Sakai, K.; Lee, S.-H.; Sato, H. The Inhibitory Components from Artocarpus incisus on Melanin Biosynthesis. Planta Med. 1998, 64, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Likhitwitayawuid, K.; Sritularak, B. A new dimeric stilbene with tyrosinase inhibitiory activity from Artocarpus gomezianus. J. Nat. Prod. 2001, 64, 1457–1459. [Google Scholar] [CrossRef]
- Ohguchi, K.; Tanaka, T.; Ito, T.; Iinuma, M.; Matsumoto, K.; Akao, Y.; Nozawa, Y. Inhibitory effects of resveratrol derivatives from dipterocarpaceae plants on tyrosinase activity. Biosci. Biotechnol. Biochem. 2003, 67, 1587–1589. [Google Scholar] [CrossRef] [Green Version]
- Likhitwitayawuid, K.; Sornsute, A.; Sritularak, B.; Ploypradith, P. Chemical transformations of oxyresveratrol (trans-2,4,3′,5′-tetrahydroxystilbene) into a potent tyrosinase inhibitor and a strong cytotoxic agent. Bioorgan. Med. Chem. Lett. 2006, 16, 5650–5653. [Google Scholar] [CrossRef]
- Burdock, G.A.; Soni, M.G.; Carabin, I.G. Evaluation of health aspects of kojic acid in food. Regul. Toxicol. Pharmacol. 2001, 33, 80–101. [Google Scholar] [CrossRef]
- Bentley, R. From miso, saké and shoyu to cosmetics: A century of science for kojic acid. Nat. Prod. Rep. 2006, 23, 1046–1062. [Google Scholar] [CrossRef]
- Kim, Y.-J.J.; Uyama, H. Tyrosinase inhibitors from natural and synthetic sources: Structure, inhibition mechanism and perspective for the future. Cell. Mol. Life Sci. 2005, 62, 1707–1723. [Google Scholar] [CrossRef]
- Kahn, V.; Ben-Shalom, N.; Zakin, V. Effect of Kojic Acid on the Oxidation of N-Acetyldopamine by Mushroom Tyrosinase. J. Agric. Food Chem. 1997, 45, 4460–4465. [Google Scholar] [CrossRef]
- Kim, H.; Choi, J.; Cho, J.K.; Kim, S.Y.; Lee, Y.-S. Solid-phase synthesis of kojic acid-tripeptides and their tyrosinase inhibitory activity, storage stability, and toxicity. Bioorgan. Med. Chem. Lett. 2004, 14, 2843–2846. [Google Scholar] [CrossRef]
- Choi, S.; Lee, S.-K.; Kim, J.-E.; Chung, M.-H.; Park, Y.-I. Aloesin inhibits hyperpigmentation induced by UV radiation. Clin. Exp. Dermatol. 2002, 27, 513–515. [Google Scholar] [CrossRef]
- Masamoto, Y.; Ando, H.; Murata, Y.; Shimoishi, Y.; Tada, M.; Takahata, K. Mushroom Tyrosinase Inhibitory Activity of Esculetin Isolated from Seeds of Euphorbia lathyris L. Biosci. Biotechnol. Biochem. 2003, 67, 631–634. [Google Scholar] [CrossRef]
- Piao, X.L.; Baek, S.H.; Park, M.K.; Park, J.H. Tyrosinase-inhibitory furanocoumarin from Angelica dahurica. Biol. Pharm. Bull. 2004, 27, 1144–1146. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Wakamatsu, K. A convenient screening method to differentiate phenolic skin whitening tyrosinase inhibitors from leukoderma-inducing phenols. J. Dermatol. Sci. 2015, 80, 18–24. [Google Scholar] [CrossRef]
- Lim, J.Y.; Ishiguro, K.; Kubo, I. Tyrosinase inhibitory p-coumaric acid from ginseng leaves. Phytother. Res. 1999, 13, 371–375. [Google Scholar] [CrossRef]
- Maghsoudi, S.; Adibi, H.; Hamzeh, M.; Ashrafi-Kooshk, M.R.; Rezaei-Tavirani, M.; Khodarahmi, R. Kinetic of Mushroom Tyrosinase Inhibition by Benzaldehyde Derivatives. J. Rep. Pharm. Sci. Med. Sci. 2013, 2, 156–164. [Google Scholar]
- Duckworth, H.W.; Coleman, J.E. Physicochemical and kinetic properties of mushroom tyrosinase. J. Biol. Chem. 1970, 245, 1613–1625. [Google Scholar] [CrossRef]
- Kubo, I.; Chen, Q.-X.; Nihei, K.-I.; Calderón, J.S.; Céspedes, C.L. Tyrosinase inhibition kinetics of anisic acid. Z. Naturforsch. C 2003, 58, 713–718. [Google Scholar] [CrossRef]
- Ha, T.J.; Tamura, S.; Kubo, I. Effects of mushroom tyrosinase on anisaldehyde. J. Agric. Food Chem. 2005, 53, 7024–7028. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, Q.; Wang, Q.; Song, K.; Qiu, L. Inhibitory effects of cinnamic acid and its derivatives on the diphenolase activity of mushroom (Agaricus bisporus) tyrosinase. Food Chem. 2005, 92, 707–712. [Google Scholar] [CrossRef]
- Lee, H.-S. Tyrosinase inhibitors of Pulsatilla cernua root-derived materials. J. Agric. Food Chem. 2002, 50, 1400–1403. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, M.; Oshima, T.; Koshio, K.; Itsuzaki, Y.; Anzai, J. Tyrosinase inhibitor from black rice bran. J. Agric. Food Chem. 2003, 51, 6953–6956. [Google Scholar] [CrossRef] [PubMed]
- Isao, K.; Kinst-Hori, I. Tyrosinase Inhibitors from Cumin. J. Agric. Food Chem. 1998, 46, 5338–5341. [Google Scholar] [CrossRef]
- Conrad, J.S.; Dawso, S.R.; Hubbard, E.R.; Meyers, T.E.; Strothkamp, K.G. Inhibitor binding to the binuclear active site of tyrosinase: Temperature, pH, and solvent deuterium isotope effects. Biochemistry 1994, 33, 5739–5744. [Google Scholar] [CrossRef]
- Zhu, J.; Yan, G.; Xu, Z.; Hu, X.; Wang, G.; Wang, T.; Zhu, W.; Hou, A.; Wang, H. Inhibitory Effects of (2′R)-2′,3′-dihydro-2′-(1-hydroxy-1-methylethyl)-2,6′-bibenzofuran-6,4′-diol on Mushroom Tyrosinase and Melanogenesis in B16-F10 Melanoma Cells. Phytother. Res. 2015, 1045, 1040–1045. [Google Scholar] [CrossRef]
- Kim, Y.M.; Yun, J.; Lee, C.-K.; Lee, H.; Min, K.R.; Kim, Y. Oxyresveratrol and Hydroxystilbene Compounds. J. Biol. Chem. 2002, 277, 16340–16344. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.S.; Wei, C.I.; Marshall, M.R. Inhibition mechanism of kojic acid on polyphenol oxidase. J. Agric. Food Chem. 1991, 39, 1897–1901. [Google Scholar] [CrossRef]
- Rho, H.S.; Goh, M.; Lee, J.K.; Ahn, S.M.; Yeon, J.H.; Yoo, D.S.; Kim, D.H.; Kim, H.G.; Cho, J.Y. Ester Derivatives of Kojic Acid and Polyphenols Containing Adamantane Moiety with Tyrosinase Inhibitory and Anti-Inflammatory Properties. Bull. Korean Chem. Soc. 2011, 32, 1411–1414. [Google Scholar] [CrossRef]
- Jeon, H.J.; Noda, M.; Maruyama, M.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Identification and Kinetic Study of Tyrosinase Inhibitors Found in Sake Lees. J. Agric. Food Chem. 2006, 54, 9827–9833. [Google Scholar] [CrossRef]
- Magid, A.A.; Voutguenne-Nazabadioko, L.; Bontemps, G.; Litaudon, M.; Lavaud, C. Tyrosinase Inhibitors and Sesquiterpene Diglycosides from Guioa Villosa. Planta Med. 2008, 74, 55–60. [Google Scholar] [CrossRef]
- Masuda, T.; Odaka, Y.; Ogawa, N.; Nakamoto, K.; Kuninaga, H. Identification of geranic acid, a tyrosinase inhibitor in lemongrass (Cymbopogon citratus). J. Agric. Food Chem. 2008, 56, 597–601. [Google Scholar] [CrossRef]
- Sabudak, T.; Khan, H.T.M.; Choudhary, M.I.; Oksuz, S. Potent Tyrosinase Inhibitors from Trifolium Balansae. Nat. Prod. Res. 2006, 20, 665–670. [Google Scholar] [CrossRef]
- Khan, M.T.H.; Khan, S.B.; Ather, A. Tyrosinase inhibitory cycloartane type triterpenoids from the methanol extract of the whole plant of Amberboa ramosa Jafri and their structure-activity relationship. Bioorgan. Med. Chem. 2006, 14, 938–943. [Google Scholar] [CrossRef]
- Ullah, F.; Hussain, H.; Hussain, J.; Bukhari, I.A.; Khan, M.T.H.; Choudhary, M.I.; Gilani, A.H.; Ahmad, V.U. Tyrosinase inhibitory pentacyclic triterpenes and analgesic and spasmolytic activities of methanol extracts of Rhododendron collettianum. Phytother. Res. 2007, 21, 1076–1081. [Google Scholar] [CrossRef]
- Devkota, K.P.; Khan, M.T.H.; Ranjit, R.; Meli Lannang, A.; Samreen; Iqbal Choudhary, M. Tyrosinase inhibitory and antileishmanial constituents from the rhizomes of Paris polyphylla. Nat. Prod. Res. 2007, 21, 321–327. [Google Scholar] [CrossRef]
- Azhar-Ul-Haq; Malik, A.; Khan, M.T.H.; Anwar-Ul-Haq, A.; Khan, S.B.; Ahmad, A.; Choudhary, M.I. Tyrosinase inhibitory lignans from the methanol extract of the roots of Vitex negundo Linn. and their structure-activity relationship. Phytomedicine 2006, 13, 255–260. [Google Scholar] [CrossRef]
- Lee, S.; Kim, M.; Lee, S.; Ahn, Y.; Lee, H.; Lee, S.E.; Kim, M.K. Inhibitory effects of Cinnamomum cassia bark-derived materials on mushroom tyrosinase. Food Sci. Biotechnol. 2000, 9, 330–333. [Google Scholar]
- Kubo, I.; Yokokawa, Y. Two tyrosinase inhibiting flavonol glycosides from Buddleia coriacea. Phytochemistry 1992, 31, 1075–1077. [Google Scholar] [CrossRef]
- Kubo, I.; Kinst-Hori, I. 2-Hydroxy-4-methoxybenzaldehyde: A potent tyrosinase inhibitor from African medicinal plants. Planta Med. 1999, 65, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Ya, W.; Chun-Meng, Z.; Tao, G.; Yi-Lin, Z.; Ping, Z. Preliminary screening of 44 plant extracts for anti-tyrosinase and antioxidant activities. Pak. J. Pharm. Sci. 2015, 28, 1737–1744. [Google Scholar] [PubMed]
- Kubo, I.; Kinst-Hori, I. Flavonols from saffron flower: Tyrosinase inhibitory activity and inhibition mechanism. J. Agric. Food Chem. 1999, 47, 4121–4125. [Google Scholar] [CrossRef] [PubMed]
- Brewer, M.S. Natural Antioxidants: Sources, Compounds, Mechanisms of Action, and Potential Applications. Compr. Rev. Food Sci. Food Saf. 2011, 10, 221–247. [Google Scholar] [CrossRef]
- Chai, W.M.; Wei, Q.M.; Deng, W.L.; Zheng, Y.L.; Chen, X.Y.; Huang, Q.; Ou-Yang, C.; Peng, Y.Y. Anti-melanogenesis properties of condensed tannins from: Vigna angularis seeds with potent antioxidant and DNA damage protection activities. Food Funct. 2019, 10, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Biswas, R.; Chanda, J.; Kar, A.; Mukherjee, P.K. Tyrosinase inhibitory mechanism of betulinic acid from Dillenia indica. Food Chem. 2017, 232, 689–696. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, X.; Ni, H.; Du, X.; Chen, F.; Jiang, Z.; Li, Q. Identification and Characterization of the Tyrosinase Inhibitory Activity of Caffeine from Camellia Pollen. J. Agric. Food Chem. 2019, 67, 12741–12751. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Leem, H.H.; Lee, G.Y. The guanidine pseudoalkaloids 10-methoxy- leonurine and leonurine act as competitive inhibitors of tyrosinase. Biomolecules 2020, 10, 174. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Li, J.; Liu, Z.; Zheng, M.; Ge, H.; Xu, J. Enhancing Molecular Shape Comparison by Weighted Gaussian Functions. J. Chem. Inf. Modeling 2013, 53, 1967–1978. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, H.; Li, H. SHAFTS: A Hybrid Approach for 3D Molecular Similarity Calculation. 1. Method and Assessment of Virtual Screening. J. Chem. Inf. Model. 2011, 51, 2372–2385. [Google Scholar] [CrossRef]
- Halgren, T.A. Potential energy functions. Curr. Opin. Struct. Biol. 1995, 5, 205–210. [Google Scholar] [CrossRef]
- Case, D.; Cerutti, D.S.; Cheatham, T.; Darden, T.; Duke, R.; Giese, T.J.; Gohlke, H.; Götz, A.; Greene, D.; Homeyer, N.; et al. Amber 2017; University of California: San Francisco, CA, USA, 2017. [Google Scholar] [CrossRef]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 11, 572–584. [Google Scholar] [CrossRef]
- Liu, X.; Bai, F.; Ouyang, S.; Wang, X.; Li, H.; Jiang, H. Cyndi: A multi-objective evolution algorithm based method for bioactive molecular conformational generation. BMC Bioinform. 2009, 14, 101. [Google Scholar] [CrossRef] [Green Version]
- Cregge, R.J.; Durham, S.L.; Farr, R.A.; Gallion, S.L.; Hare, C.M.; Hoffman, R.V.; Janusz, M.J.; Kim, H.O.; Koehl, J.R.; Mehdi, S.; et al. Inhibition of human neutrophil elastase. 4. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of P2-modified, orally active peptidyl pentafluoroethyl ketones. J. Med. Chem. 1998, 41, 2461–2480. [Google Scholar] [CrossRef]
- Sendovski, M.; Kanteev, M.; Ben-Yosef, V.S.; Adir, N.; Fishman, A. First structures of an active bacterial tyrosinase reveal copper plasticity. J. Mol. Biol. 2011, 405, 227–237. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [Green Version]
- Flanagan, D. JavaScript: The Definitive Guide; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2006. [Google Scholar]
- De Volder, K.; Query, J. A generic code browser with a declarative configuration language. In International Symposium on Practical Aspects of Declarative Languages; Springer: Berlin/Heidelberg, Germany, 2006; pp. 88–102. [Google Scholar]
- Welling, L.; Thomson, L. PHP and MySQL Web Development; Sams Publishing: Carmel, Indiana, 2003. [Google Scholar]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Watts, K.S.; Dalal, P.; Murphy, R.B.; Sherman, W.; Friesner, R.A.; Shelley, J.C. ConfGen: A conformational search method for efficient generation of bioactive conformers. J. Chem. Inf. Model. 2010, 50, 534–546. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaemmanouil, C.D.; Peña-García, J.; Banegas-Luna, A.J.; Kostagianni, A.D.; Gerothanassis, I.P.; Pérez-Sánchez, H.; Tzakos, A.G. ANTIAGE-DB: A Database and Server for the Prediction of Anti-Aging Compounds Targeting Elastase, Hyaluronidase, and Tyrosinase. Antioxidants 2022, 11, 2268. https://doi.org/10.3390/antiox11112268
Papaemmanouil CD, Peña-García J, Banegas-Luna AJ, Kostagianni AD, Gerothanassis IP, Pérez-Sánchez H, Tzakos AG. ANTIAGE-DB: A Database and Server for the Prediction of Anti-Aging Compounds Targeting Elastase, Hyaluronidase, and Tyrosinase. Antioxidants. 2022; 11(11):2268. https://doi.org/10.3390/antiox11112268
Chicago/Turabian StylePapaemmanouil, Christina D., Jorge Peña-García, Antonio Jesús Banegas-Luna, Androniki D. Kostagianni, Ioannis P. Gerothanassis, Horacio Pérez-Sánchez, and Andreas G. Tzakos. 2022. "ANTIAGE-DB: A Database and Server for the Prediction of Anti-Aging Compounds Targeting Elastase, Hyaluronidase, and Tyrosinase" Antioxidants 11, no. 11: 2268. https://doi.org/10.3390/antiox11112268