N-Heterocyclic Carbene-Gold(I) Complexes Targeting Actin Polymerization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Docking Studies
2.3. Immunofluorescence Analysis
2.4. Actin Polymerization/Depolymerization Assay
3. Results
3.1. AuL4 and AuL7 Induce Dramatic Cancer Cells Morphology Changes
3.2. Docking Studies
3.3. AuL4 and AuL7 Interfere with the Normal Intracellular Actin Organization
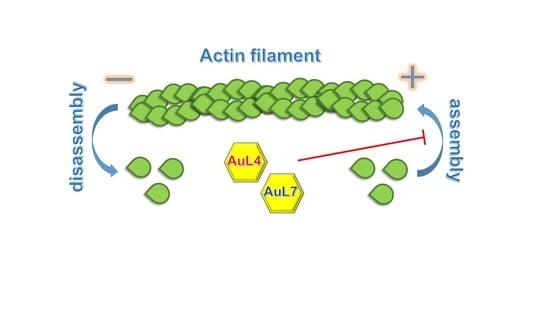
3.4. AuL4 and AuL7 Block the In Vitro Actin Polymerization Reaction but Do Not Accelerate the Depolymerization Reaction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jakupec, M.A.; Galanski, M.; Arion, V.B.; Hartinger, C.G.; Keppler, B.K. Antitumour metal compounds: More than theme and variations. Dalton Trans. 2008, 183–194. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 44, 4565–4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, B.; Casini, A. A golden future in medicinal inorganic chemistry: The promise of anticancer gold organometallic compounds. Dalton Trans. 2014, 43, 4209–4219. [Google Scholar] [CrossRef] [PubMed]
- Gaynor, D.; Griffith, D.M. The prevalence of metal-based drugs as therapeutic or diagnostic agents: Beyond platinum. Dalton Trans. 2012, 41, 13239–13257. [Google Scholar] [CrossRef]
- Orvig, C.; Abrams, M.J. Medicinal inorganic chemistry: Introduction. Chem. Rev. 1999, 99, 2201–2204. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, N.L. Comparative pharmacokinetics of parenteral and oral gold compounds. J. Rheumatol. Suppl. 1982, 8, 99–109. [Google Scholar]
- Hindi, K.M.; Panzner, M.J.; Tessier, C.A.; Cannon, C.L.; Youngs, W.J. The medicinal applications of imidazolium carbene-metal complexes. Chem. Rev. 2009, 109, 3859–3884. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Gust, R. Metal N-heterocyclic carbene complexes as potential antitumor metallodrugs. Chem. Soc. Rev. 2013, 42, 755–773. [Google Scholar] [CrossRef]
- Ceramella, J.; Mariconda, A.; Iacopetta, D.; Saturnino, C.; Barbarossa, A.; Caruso, A.; Rosano, C.; Sinicropi, M.S.; Longo, P. From coins to cancer therapy: Gold, silver and copper complexes targeting human topoisomerases. Bioorg. Med. Chem. Lett. 2020, 30, 126905. [Google Scholar] [CrossRef]
- Hickey, J.L.; Ruhayel, R.A.; Barnard, P.J.; Baker, M.V.; Berners-Price, S.J.; Filipovska, A. Mitochondria-targeted chemotherapeutics: The rational design of gold(I) N-heterocyclic carbene complexes that are selectively toxic to cancer cells and target protein selenols in preference to thiols. J. Am. Chem. Soc. 2008, 130, 12570–12571. [Google Scholar] [CrossRef] [PubMed]
- Barnard, P.J.; Berners-Price, S.J. Targeting the mitochondrial cell death pathway with gold compounds. Coord. Chem. Rev. 2007, 251, 1889–1902. [Google Scholar] [CrossRef]
- Krishnamurthy, D.; Karver, M.R.; Fiorillo, E.; Orru, V.; Stanford, S.M.; Bottini, N.; Barrios, A.M. Gold(I)-mediated inhibition of protein tyrosine phosphatases: A detailed in vitro and cellular study. J. Med. Chem. 2008, 51, 4790–4795. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.K.; Zou, T.; Cao, B.; Lee, P.Y.; Fung, Y.M.; Hu, D.; Lok, C.N.; Che, C.M. Cyclometalated Gold(III) Complexes Containing N-Heterocyclic Carbene Ligands Engage Multiple Anti-Cancer Molecular Targets. Angew. Chem. Int. Ed. Engl. 2017, 56, 3892–3896. [Google Scholar] [CrossRef] [PubMed]
- Iacopetta, D.; Rosano, C.; Sirignano, M.; Mariconda, A.; Ceramella, J.; Ponassi, M.; Saturnino, C.; Sinicropi, M.S.; Longo, P. Is the Way to Fight Cancer Paved with Gold? Metal-Based Carbene Complexes with Multiple and Fascinating Biological Features. Pharmaceuticals 2020, 13, 91. [Google Scholar] [CrossRef]
- Magherini, F.; Fiaschi, T.; Valocchia, E.; Becatti, M.; Pratesi, A.; Marzo, T.; Massai, L.; Gabbiani, C.; Landini, I.; Nobili, S.; et al. Antiproliferative effects of two gold(I)-N-heterocyclic carbene complexes in A2780 human ovarian cancer cells: A comparative proteomic study. Oncotarget 2018, 9, 28042–28068. [Google Scholar] [CrossRef]
- Muenzner, J.K.; Biersack, B.; Albrecht, A.; Rehm, T.; Lacher, U.; Milius, W.; Casini, A.; Zhang, J.J.; Ott, I.; Brabec, V.; et al. Ferrocenyl-Coupled N-Heterocyclic Carbene Complexes of Gold(I): A Successful Approach to Multinuclear Anticancer Drugs. Chemistry 2016, 22, 18953–18962. [Google Scholar] [CrossRef]
- Iacopetta, D.; Mariconda, A.; Saturnino, C.; Caruso, A.; Palma, G.; Ceramella, J.; Muia, N.; Perri, M.; Sinicropi, M.S.; Caroleo, M.C.; et al. Novel Gold and Silver Carbene Complexes Exert Antitumor Effects Triggering the Reactive Oxygen Species Dependent Intrinsic Apoptotic Pathway. ChemMedChem 2017, 12, 2054–2065. [Google Scholar] [CrossRef]
- Dhamodharan, R.; Jordan, M.A.; Thrower, D.; Wilson, L.; Wadsworth, P. Vinblastine suppresses dynamics of individual microtubules in living interphase cells. Mol. Biol. Cell 1995, 6, 1215–1229. [Google Scholar] [CrossRef] [Green Version]
- Rizza, P.; Pellegrino, M.; Caruso, A.; Iacopetta, D.; Sinicropi, M.S.; Rault, S.; Lancelot, J.C.; El-Kashef, H.; Lesnard, A.; Rochais, C.; et al. 3-(Dipropylamino)-5-hydroxybenzofuro[2,3-f]quinazolin-1(2H)-one (DPA-HBFQ-1) plays an inhibitory role on breast cancer cell growth and progression. Eur. J. Med. Chem. 2016, 107, 275–287. [Google Scholar] [CrossRef]
- Ceramella, J.; Caruso, A.; Occhiuzzi, M.A.; Iacopetta, D.; Barbarossa, A.; Rizzuti, B.; Dallemagne, P.; Rault, S.; El-Kashef, H.; Saturnino, C.; et al. Benzothienoquinazolinones as new multi-target scaffolds: Dual inhibition of human Topoisomerase I and tubulin polymerization. Eur. J. Med. Chem. 2019, 181, 111583. [Google Scholar] [CrossRef] [PubMed]
- Rebowski, G.; Boczkowska, M.; Drazic, A.; Ree, R.; Goris, M.; Arnesen, T.; Dominguez, R. Mechanism of actin N-terminal acetylation. Sci. Adv. 2020, 6, eaay8793. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanner, M.F.; Duncan, B.S.; Carrillo, C.J.; Olson, A.J. Integrating computation and visualization for biomolecular analysis: An example using python and AVS. Pac. Symp. Biocomput. 1999, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Cesarini, S.; Spallarossa, A.; Ranise, A.; Schenone, S.; Rosano, C.; La Colla, P.; Sanna, G.; Busonera, B.; Loddo, R. N-acylated and N,N’-diacylated imidazolidine-2-thione derivatives and N,N’-diacylated tetrahydropyrimidine-2(1H)-thione analogues: Synthesis and antiproliferative activity. Eur. J. Med. Chem. 2009, 44, 1106–1118. [Google Scholar] [CrossRef]
- Rosano, C.; Hunt, M.J.O.; Brach, J.; Newman, A.B.; Studenski, S.; Verghese, J.; Weissfeld, L. Brain Anatomical Correlates of Gait Variability in High Functioning Older Adults: Repeatability across Two Independent Studies (Cns). Gerontologist 2012, 52, 319–320. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Ceramella, J.; Loizzo, M.R.; Iacopetta, D.; Bonesi, M.; Sicari, V.; Pellicano, T.M.; Saturnino, C.; Malzert-Freon, A.; Tundis, R.; Sinicropi, M.S. Anchusa azurea Mill. (Boraginaceae) aerial parts methanol extract interfering with cytoskeleton organization induces programmed cancer cells death. Food Funct. 2019, 10, 4280–4290. [Google Scholar] [CrossRef] [PubMed]
- Lappano, R.; Rosano, C.; Pisano, A.; Santolla, M.F.; De Francesco, E.M.; De Marco, P.; Dolce, V.; Ponassi, M.; Felli, L.; Cafeo, G.; et al. A calixpyrrole derivative acts as an antagonist to GPER, a G-protein coupled receptor: Mechanisms and models. Dis. Model Mech. 2015, 8, 1237–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinicropi, M.S.; Lappano, R.; Caruso, A.; Santolla, M.F.; Pisano, A.; Rosano, C.; Capasso, A.; Panno, A.; Lancelot, J.C.; Rault, S.; et al. (6-bromo-1,4-dimethyl-9H-carbazol-3-yl-methylene)-hydrazine (carbhydraz) acts as a GPER agonist in breast cancer cells. Curr. Top. Med. Chem. 2015, 15, 1035–1042. [Google Scholar] [CrossRef]
- Stec-Martyna, E.; Ponassi, M.; Miele, M.; Parodi, S.; Felli, L.; Rosano, C. Structural comparison of the interaction of tubulin with various ligands affecting microtubule dynamics. Curr. Cancer Drug Targets 2012, 12, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Maji, M.; Ruturaj; Purkait, K.; Gupta, A.; Mukherjee, A. Synthesis, Structure, Stability, and Inhibition of Tubulin Polymerization by Ru(II)-p-Cymene Complexes of Trimethoxyaniline-Based Schiff Bases. Inorg. Chem. 2019, 58, 9213–9224. [Google Scholar] [CrossRef] [PubMed]
- Ceresa, C.; Nicolini, G.; Semperboni, S.; Gandin, V.; Monfrini, M.; Avezza, F.; Alberti, P.; Bravin, A.; Pellei, M.; Santini, C.; et al. Evaluation of the Profile and Mechanism of Neurotoxicity of Water-Soluble [Cu(P)4]PF6 and [Au(P)4]PF6 (P = thp or PTA) Anticancer Complexes. Neurotox Res. 2018, 34, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Kaps, L.; Biersack, B.; Muller-Bunz, H.; Mahal, K.; Munzner, J.; Tacke, M.; Mueller, T.; Schobert, R. Gold(I)-NHC complexes of antitumoral diarylimidazoles: Structures, cellular uptake routes and anticancer activities. J. Inorg. Biochem. 2012, 106, 52–58. [Google Scholar] [CrossRef]
- Khanna, S.; Jana, B.; Saha, A.; Kurkute, P.; Ghosh, S.; Verma, S. Targeting cytotoxicity and tubulin polymerization by metal-carbene complexes on a purine tautomer platform. Dalton Trans. 2014, 43, 9838–9842. [Google Scholar] [CrossRef]
- Muenzner, J.K.; Biersack, B.; Kalie, H.; Andronache, I.C.; Kaps, L.; Schuppan, D.; Sasse, F.; Schobert, R. Gold(I) biscarbene complexes derived from vascular-disrupting combretastatin A-4 address different targets and show antimetastatic potential. ChemMedChem 2014, 9, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Haeberle, S.; Shytaj, I.L.; Gama-Brambila, R.A.; Theobald, J.; Ghafoory, S.; Wolker, J.; Basu, U.; Schmidt, C.; Timm, A.; et al. NHC-gold compounds mediate immune suppression through induction of AHR-TGFbeta1 signalling in vitro and in scurfy mice. Commun. Biol. 2020, 3, 10. [Google Scholar] [CrossRef]
- Pasquier, E.; Kavallaris, M. Microtubules: A dynamic target in cancer therapy. IUBMB Life 2008, 60, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Fenteany, G.; Zhu, S. Small-molecule inhibitors of actin dynamics and cell motility. Curr. Top. Med. Chem. 2003, 3, 593–616. [Google Scholar] [CrossRef]
- Aseervatham, J. Cytoskeletal Remodeling in Cancer. Biology 2020, 9, 385. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 2007, 1773, 642–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenblum, M.D.; Shivers, R.R. ‘Rings’ of F-actin form around the nucleus in cultured human MCF7 adenocarcinoma cells upon exposure to both taxol and taxotere. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 2000, 125, 121–131. [Google Scholar] [CrossRef]
- Foissner, I.; Wasteneys, G.O. Wide-ranging effects of eight cytochalasins and latrunculin A and B on intracellular motility and actin filament reorganization in characean internodal cells. Plant Cell Physiol. 2007, 48, 585–597. [Google Scholar] [CrossRef]
- Fujiwara, I.; Zweifel, M.E.; Courtemanche, N.; Pollard, T.D. Latrunculin A Accelerates Actin Filament Depolymerization in Addition to Sequestering Actin Monomers. Curr. Biol. 2018, 28, 3183–3192. [Google Scholar] [CrossRef] [Green Version]
- Danowski, B.A. Fibroblast contractility and actin organization are stimulated by microtubule inhibitors. J. Cell Sci. 1989, 93 Pt 2, 255–266. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacopetta, D.; Ceramella, J.; Rosano, C.; Mariconda, A.; Pellegrino, M.; Sirignano, M.; Saturnino, C.; Catalano, A.; Aquaro, S.; Longo, P.; et al. N-Heterocyclic Carbene-Gold(I) Complexes Targeting Actin Polymerization. Appl. Sci. 2021, 11, 5626. https://doi.org/10.3390/app11125626
Iacopetta D, Ceramella J, Rosano C, Mariconda A, Pellegrino M, Sirignano M, Saturnino C, Catalano A, Aquaro S, Longo P, et al. N-Heterocyclic Carbene-Gold(I) Complexes Targeting Actin Polymerization. Applied Sciences. 2021; 11(12):5626. https://doi.org/10.3390/app11125626
Chicago/Turabian StyleIacopetta, Domenico, Jessica Ceramella, Camillo Rosano, Annaluisa Mariconda, Michele Pellegrino, Marco Sirignano, Carmela Saturnino, Alessia Catalano, Stefano Aquaro, Pasquale Longo, and et al. 2021. "N-Heterocyclic Carbene-Gold(I) Complexes Targeting Actin Polymerization" Applied Sciences 11, no. 12: 5626. https://doi.org/10.3390/app11125626