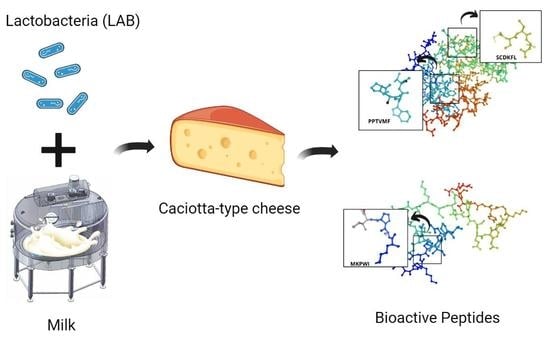
Effect of Lactobacteria on Bioactive Peptides and Their Sequence Identification in Mature Cheese
Abstract
:1. Introduction
2. Materials and Methods
2.1. Model Caciotta-Type Cheese Production
| Starting Components | Basic Cheese-Making Parameters | ||
|---|---|---|---|
| cow milk | 10 L | Pasteurization milk | 64 °C, 30 min |
| annatto (for winter milk) * | 5–6 drops | Inoculation (fermentation) | 34 °C, 10 min |
| * prefabricated bacterial mesophilic culture (Lc. lactis subsp. lactis V-1568; Lc. lactis subsp. cremoris V-1569) (control cheese A) (National Bioresource Center Russian Collection of Industrial Microorganisms (VKPM)) | 150 g (0.5% of the milk volume | Flocculation multiplier | 3 |
| ** prefabricated bacterial mesophilic culture (Lc. lactis subsp. lactis V-1568; Lc. lactis subsp. cremoris V-1569; Lac. casei V-9227) (experimental cheese B) (National Bioresource Center Russian Collection of Industrial Microorganisms (VKPM)) | 100 g (50/50, respectively) | Cheese cube size | 1.0 cm |
| calcium chloride (10–20% solution CaCl2) in an aqueous solution (for pasteurized and winter milk) | 10 mL | Granular curd heating temperature | 45 °C |
| milk-clotting enzyme (“Carlina” (composition: 90% rennet chymosin, 10% pepsin; manufacturer: Danisco France SAS, France) | 0.35 g (in the amount necessary for 12–15 min of flocculation time) | Stuffature (the cheese head was turned eight times); | 1.5 h at 45 °C |
| Development at room temperature ≥ 22 °C | 4 h | ||
| Maturation time (see final product in Figure 1). | 90 days | ||

2.2. Micrographs of the Lactobacilli
- – Type of measurement—according to the Fraunhofer method;
- – Measurement range—from 0.1 μm to 1021.87 μm;
- – Resolution—102 channels;
- – Absorption—10.00%;
- – Measurement duration—90 scans.
2.3. Protein Analyses
2.4. The Molecular Weight Distribution
2.5. Amino Acid Analyses
2.6. Biological Activity of Peptides In Silico
2.7. Visualization of Dihedral Amino-Acid Angles
2.8. D protein Structure Modeling
2.9. Modeling the Structure of Peptides
2.10. Hydrophobicity and Hydrophilicity of Proteins
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hayaloglu, A.A.; Brechany, E.Y. Influence of Milk Pasteurization and Scalding Temperature on the Volatile Compounds of Malatya, a Farmhouse Halloumi-Type Cheese. Lait 2007, 87, 39–57. [Google Scholar] [CrossRef] [Green Version]
- Prosekov, A.Y.; Ivanova, S.A. Food security: The challenge of the present. Geoforum 2018, 91, 73–77. [Google Scholar] [CrossRef]
- Vesnina, A.; Prosekov, A.; Kozlova, O.; Atuchin, V. Genes and Eating Preferences, Their Roles in Personalized Nutrition. Genes 2020, 11, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Lista, J.; Alcala-Diaz, J.F.; Torres-Peña, J.D.; Quintana-Navarro, G.M.; Fuentes, F.; Garcia-Rios, A.; Ortiz-Morales, A.M.; Gonzalez-Requero, A.I.; Perez-Caballero, A.I.; Yubero-Serrano, E.M.; et al. Long-Term Secondary Prevention of Cardiovascular Disease with a Mediterranean Diet and a Low-Fat Diet (CORDIOPREV): A Randomised Controlled Trial. Lancet 2022, 399, 1876–1885. [Google Scholar] [CrossRef]
- Vargas-Rodríguez, I.; Reyes-Castro, L.A.; Pacheco-López, G.; Lomas-Soria, C.; Zambrano, E.; Díaz-Ruíz, A.; Diaz-Cintra, S. Postnatal Exposure to Lipopolysaccharide Combined with High-Fat Diet Consumption Induces Immune Tolerance without Prevention in Spatial Working Memory Impairment. Behav. Brain Res. 2022, 423, 113776. [Google Scholar] [CrossRef]
- Nabuuma, D.; Reimers, C.; Hoang, K.T.; Stomph, T.; Swaans, K.; Raneri, J.E. Impact of Seed System Interventions on Food and Nutrition Security in Low- and Middle-Income Countries: A Scoping Review. Glob. Food Secur. 2022, 33, 100638. [Google Scholar] [CrossRef]
- Vesnina, A.; Prosekov, A.; Atuchin, V.; Minina, V.; Ponasenko, A. Tackling atherosclerosis via selected nutrition. Int. J. Mol. Sci. 2022, 23, 8233. [Google Scholar] [CrossRef]
- Santillán-Urquiza, E.; Méndez-Rojas, M.Á.; Vélez-Ruiz, J.F. Fortification of Yogurt with Nano and Micro Sized Calcium, Iron and Zinc, Effect on the Physicochemical and Rheological Properties. LWT-Food Sci. Technol. 2017, 80, 462–469. [Google Scholar] [CrossRef]
- Talbot-Walsh, G.; Kannar, D.; Selomulya, C. A Review on Technological Parameters and Recent Advances in the Fortification of Processed Cheese. Trends Food Sci. Technol. 2018, 81, 193–202. [Google Scholar] [CrossRef]
- Artyukhova, S.; Kozlova, O.; Tolstoguzova, T. Developing freeze-dried bioproducts for the Russian military in the Arctic. Foods Raw Mater. 2019, 7, 202–209. [Google Scholar] [CrossRef]
- Kolbina, A.Y.; Ulrikh, E.V.; Voroshilin, R.A. Analysis of consumer motivations of the Kemerovo city residents in relation to functional food products. EurAsian J. BioSci. 2020, 14, 6365–6369. [Google Scholar]
- Rivero-Pino, F.; Espejo-Carpio, F.J.; Guadix, E.M. Evaluation of the Bioactive Potential of Foods Fortified with Fish Protein Hydrolysates. Food Res. Int. 2020, 137, 109572. [Google Scholar] [CrossRef] [PubMed]
- Milentyeva, I.; Le, V.; Kozlova, O.; Velichkovich, N.; Fedorova, A.; Loseva, A.; Yustratov, V. Secondary metabolites in in vitro cultures of Siberian medicinal plants: Content, antioxidant properties, and antimicrobial characteristics. Foods Raw Mater. 2021, 9, 153–163. [Google Scholar] [CrossRef]
- Prosekov, A.Y.; Altshuler, O.G.; Kurbanova, M.G. Quality and Safety of Game Meat from the Biocenosis of the Beloosipovo Mercury Deposit (part 2). Food Process. Tech. Technol. 2021, 51, 654–663. [Google Scholar] [CrossRef]
- Dary, O.; Guamuch-Castañeda, M.; Mora, J.O. Food Fortification: Technological Aspects. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Carocho, M.; Ferreira, I.C.F.R. A Review on Antioxidants, Prooxidants and Related Controversy: Natural and Synthetic Compounds, Screening and Analysis Methodologies and Future Perspectives. Food Chem. Toxicol. 2013, 51, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Geboers, S.; Stappaerts, J.; Mols, R.; Snoeys, J.; Tack, J.; Annaert, P.; Augustijns, P. The Effect of Food on the Intraluminal Behavior of Abiraterone Acetate in Man. J. Pharm. Sci. 2016, 105, 2974–2981. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Wang, X.; Zheng, Y.; Li, Y.; Guo, M.; Yan, Z. Novel Antioxidant Peptides Identified in Millet Bran Glutelin-2 Hydrolysates: Purification, in Silico Characterization and Security Prediction, and Stability Profiles under Different Food Processing Conditions. LWT-Food Sci. Technol. 2022, 164, 113634. [Google Scholar] [CrossRef]
- Sila, A.; Bougatef, A. Antioxidant Peptides from Marine By-Products: Isolation, Identification and Application in Food Systems. A Review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Vesnina, A.D.; Prosekov, A.Y.; Kozlova, O.V.; Kurbanova, M.G.; Kozlenko, E.A.; Golubtsova, Y.V. Development of a probiotic consortium for people with cancer. Vestn. VGUIT [Proc. VSUET] 2021, 83, 219–232. [Google Scholar] [CrossRef]
- Rubel, I.A.; Iraporda, C.; Manrique, G.D.; Genovese, D.B. Jerusalem Artichoke (Helianthus Tuberosus L.) Inulin as a Suitable Bioactive Ingredient to Incorporate into Spreadable Ricotta Cheese for the Delivery of Probiotic. Bioact. Carbohydr. Diet. Fibre 2022, 28, 100325. [Google Scholar] [CrossRef]
- Marco, M.D.; Baker, M.L.; Daszak, P.; Barro, P.D.; Eskew, E.A.; Godde, C.M.; Harwood, T.D.; Herrero, M.; Hoskins, A.J.; Johnson, E.; et al. Sustainable Development Must Account for Pandemic Risk. Proc. Natl. Acad. Sci. USA 2020, 117, 3888–3892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Babich, O.; Sukhikh, S.; Zimina, M.; Milentyeva, I. Antibiotic activity and resistance of lactic acid bacteria and other antagonistic bacteriocin-producing microorganisms. Foods Raw Mater. 2020, 8, 377–384. [Google Scholar] [CrossRef]
- Kosmerl, E.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Jiménez-Flores, R.; García-Cano, I. Improving Human Health with Milk Fat Globule Membrane, Lactic Acid Bacteria, and Bifidobacteria. Microorganisms 2021, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Golovach, T.N.; Kurchenko, V.P.; Zhygankov, V.G.; Evdokimov, I.A. Determination of physicochemical, immunochemical and antioxidant properties, toxicological and hygienic assessment of whey protein COMCENTRATE and its hydrolysate. Foods Raw Mater. 2015, 3, 105–114. [Google Scholar] [CrossRef]
- Moreira, G.M.M.; Costa, R.G.B.; Teodoro, V.A.M.; Paula, J.C.J.; Sobral, D.; Fernandes, C.; Gloria, M.B.A. Effect of Ripening Time on Proteolysis, Free Amino Acids, Bioactive Amines and Texture Profile of Gorgonzola-Type Cheese. LWT-Food Sci. Technol. 2018, 98, 583–590. [Google Scholar] [CrossRef]
- Hanlon, M.; Choi, J.; Goddik, L.; Park, S.H. Microbial and Chemical Composition of Cheddar Cheese Supplemented with Prebiotics from Pasteurized Milk to Aging. J. Dairy Sci. 2022, 105, 2058–2068. [Google Scholar] [CrossRef]
- Orlyuk, Y.T.; Stepanishchev, M.I. Assessment of proteolysis and lipolysis intensity in Pechersky cheese ripening in the presence of Penicillium camemberti and Penicillium roqueforti molds. Foods Raw Mater. 2014, 2, 36–39. [Google Scholar] [CrossRef]
- Asensio-Grau, A.; Peinado, I.; Heredia, A.; Andrés, A. In Vitro Study of Cheese Digestion: Effect of Type of Cheese and Intestinal Conditions on Macronutrients Digestibility. LWT-Food Sci. Technol. 2019, 113, 108278. [Google Scholar] [CrossRef]
- Villamil, R.-A.; Guzmán, M.-P.; Ojeda-Arredondo, M.; Cortés, L.Y.; Archila, E.G.; Giraldo, A.; Mondragón, A.-I. Cheese Fortification through the Incorporation of UFA-Rich Sources: A Review of Recent (2010–2020) Evidence. Heliyon 2021, 7, e05785. [Google Scholar] [CrossRef]
- Ramezani, M.; Hosseini, S.M.; Ferrocino, I.; Amoozegar, M.A.; Cocolin, L. Molecular Investigation of Bacterial Communities during the Manufacturing and Ripening of Semi-Hard Iranian Liqvan Cheese. Food Microbiol. 2017, 66, 64–71. [Google Scholar] [CrossRef]
- Mohammed, S.; Çon, A.H. Isolation and Characterization of Potential Probiotic Lactic Acid Bacteria from Traditional Cheese. LWT-Food Sci. Technol. 2021, 152, 112319. [Google Scholar] [CrossRef]
- Yang, F.; Chen, X.; Huang, M.; Yang, Q.; Cai, X.; Chen, X.; Du, M.; Huang, J.; Wang, S. Molecular Characteristics and Structure–Activity Relationships of Food-Derived Bioactive Peptides. J. Integr. Agric. 2021, 20, 2313–2332. [Google Scholar] [CrossRef]
- Kaneko, K. Appetite Regulation by Plant-Derived Bioactive Peptides for Promoting Health. Peptides 2021, 144, 170608. [Google Scholar] [CrossRef]
- Romero-Garay, M.G.; Montalvo-González, E.; Hernández-González, C.; Soto-Domínguez, A.; Becerra-Verdín, E.M.; García-Magaña, M.D.L. Bioactivity of Peptides Obtained from Poultry By-Products: A Review. Food Chem. X 2022, 13, 100181. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Lafarga, T.; Wilm, M.; Wynne, K.; Hayes, M. Bioactive Hydrolysates from Bovine Blood Globulins: Generation, Characterisation, and in Silico Prediction of Toxicity and Allergenicity. J. Funct. Foods 2016, 24, 142–155. [Google Scholar] [CrossRef] [Green Version]
- Babich, O.; Milentyeva, I.; Dyshlyuk, L.; Ostapova, E.; Altshuler, O. Structure and Properties of Antimicrobial Peptides Produced by Antagonist Microorganisms Isolated from Siberian Natural Objects. Foods Raw Mater. 2022, 10, 27–39. [Google Scholar] [CrossRef]
- Vilcacundo, R.; Martínez-Villaluenga, C.; Hernández-Ledesma, B. Release of Dipeptidyl Peptidase IV, α-Amylase and α-Glucosidase Inhibitory Peptides from Quinoa (Chenopodium Quinoa Willd.) during in Vitro Simulated Gastrointestinal Digestion. J. Funct. Foods 2017, 35, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Kehinde, B.A.; Sharma, P.; Sharma, D.; Kaur, S. Recently Isolated Food-Derived Antihypertensive Hydrolysates and Peptides: A Review. Food Chem. 2021, 346, 128719. [Google Scholar] [CrossRef] [PubMed]
- Lavelli, V.; Proserpio, C.; Gallotti, F.; Laureati, M.; Pagliarini, E. Circular Reuse of Bio-Resources: The Role of Pleurotus Spp. in the Development of Functional Foods. Food Funct. 2018, 9, 1353–1372. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.A.; Pintado, M.E. Bioactive Peptides Derived from Marine Sources: Biological and Functional Properties. Trends Food Sci. Technol. 2022, 119, 348–370. [Google Scholar] [CrossRef]
- Milentyeva, I.S.; Davydenko, N.I.; Raschepkin, A.N. Casein Proteolysis in Bioactive Peptide Production: Optimal Operating Parameters. Food Process. Tech. Technol. 2020, 4, 726–735. [Google Scholar] [CrossRef]
- Trinidad-Calderón, P.A.; Acosta-Cruz, E.; Rivero-Masante, M.N.; Díaz-Gómez, J.L.; García-Lara, S.; López-Castillo, L.M. Maize Bioactive Peptides: From Structure to Human Health. J. Cereal Sci. 2021, 100, 103232. [Google Scholar] [CrossRef]
- Shivanna, S.K.; Nataraj, B.H. Revisiting Therapeutic and Toxicological Fingerprints of Milk-Derived Bioactive Peptides: An Overview. Food Biosci. 2020, 38, 100771. [Google Scholar] [CrossRef]
- Tonolo, F.; Folda, A.; Cesaro, L.; Scalcon, V.; Marin, O.; Ferro, S.; Bindoli, A.; Rigobello, M.P. Milk-Derived Bioactive Peptides Exhibit Antioxidant Activity through the Keap1-Nrf2 Signaling Pathway. J. Funct. Foods 2020, 64, 103696. [Google Scholar] [CrossRef]
- Hafeez, Z.; Cakir-Kiefer, C.; Roux, E.; Perrin, C.; Miclo, L.; Dary-Mourot, A. Strategies of Producing Bioactive Peptides from Milk Proteins to Functionalize Fermented Milk Products. Food Res. Int. 2014, 63, 71–80. [Google Scholar] [CrossRef]
- Ayala-Niño, A.; Castañeda-Ovando, A.; Jaimez-Ordaz, J.; Rodríguez-Serrano, G.M.; Sánchez-Franco, J.A.; González-Olivares, L.G. Novel Bioactive Peptides Sequences Released Byin Vitro Digestion of Proteins Isolated From Amaranthus Hypochondriacus. Nat. Prod. Res. 2022, 36, 3485–3488. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Liu, H.; Zhang, Y.; Zhang, S.; Liu, T.; Wang, D. Review on Plant-Derived Bioactive Peptides: Biological Activities, Mechanism of Action and Utilizations in Food Development. J. Future Foods 2022, 2, 143–159. [Google Scholar] [CrossRef]
- Okoye, C.O.; Ezeorba, T.P.C.; Okeke, E.S.; Okagu, I.U. Recent Findings on the Isolation, Identification and Quantification of Bioactive Peptides. Appl. Food Res. 2022, 2, 100065. [Google Scholar] [CrossRef]
- Voroshilin, R.A.; Kurbanova, M.G.; Yustratov, V.P.; Larichev, T.A. Identifying Bioactive Peptides from Poultry By-Products. Food Process. Tech. Technol. 2022, 52, 545–554. (In Russian) [Google Scholar] [CrossRef]
- Lanza, J.G.; Churion, P.C.; Gomez, N. Comparación entre el método Kjeldahl tradicional y el método Dumas automatizado (N cube) para la determinación de proteínas en distintas clases de alimentos. Saber 2016, 28, 245–249. [Google Scholar]
- Voroshilin, R.A.; Kurbanova, M.G.; Makhambetov, E.M.; Petrov, A.N.; Khelef, M.E.A. Effect of gelatin drying methods on its amphiphilicity. Foods Raw Mater. 2022, 10, 252–261. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, S.H.; Wimley, W.C. Hydrophobic Interactions of Peptides with Membrane Interfaces. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 1998, 1376, 339–352. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Erginkaya, Z.; Konuray-Altun, G. Potential Biotherapeutic Properties of Lactic Acid Bacteria in Foods. Food Biosci. 2022, 46, 101544. [Google Scholar] [CrossRef]
- Demarigny, Y. LACTOCOCCUS|Lactococcus Lactis Subsp. Lactis and Cremoris. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 442–446. [Google Scholar] [CrossRef]
- Poudel, R.; Thunell, R.K.; Oberg, C.J.; Overbeck, S.; Lefevre, M.; Oberg, T.S.; McMahon, D.J. Comparison of Growth and Survival of Single Strains of Lactococcus Lactis and Lactococcus Cremoris during Cheddar Cheese Manufacture. J. Dairy Sci. 2022, 105, 2069–2081. [Google Scholar] [CrossRef]
- Kristo, E.; Miao, Z.; Corredig, M. The Role of Exopolysaccharide Produced by Lactococcus Lactis Subsp. Cremoris in Structure Formation and Recovery of Acid Milk Gels. Int. Dairy J. 2011, 21, 656–662. [Google Scholar] [CrossRef]
- Minervini, F.; Calasso, M. Lactobacillus Casei Group. In Encyclopedia of Dairy Sciences, 3rd ed.; McSweeney, P.L.H., McNamara, J.P., Eds.; Academic Press: Oxford, UK, 2022; pp. 275–286. [Google Scholar] [CrossRef]
- NabizadehAsl, L.; Sendur, S.N.; Ozer, B.; Lay, I.; Erbas, T.; Buyuktuncer, Z. Acute and Short-Term Effects of Lactobacillus Paracasei Subsp. Paracasei 431 and Inulin Intake on Appetite Control and Dietary Intake: A Two-Phases Randomized, Double Blind, Placebo-Controlled Study. Appetite 2022, 169, 105855. [Google Scholar] [CrossRef]
- Perna, A.; Simonetti, A.; Intaglietta, I.; Gambacorta, E. Effects of Genetic Type, Stage of Lactation, and Ripening Time on Caciocavallo Cheese Proteolysis. J. Dairy Sci. 2014, 97, 1909–1917. [Google Scholar] [CrossRef]
- Fox, P.F. Cheese: An Overview. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., Ed.; Springer: Boston, MA, USA, 1993; pp. 1–36. [Google Scholar] [CrossRef]
- Lee, H.W.; Lu, Y.; Zhang, Y.; Fu, C.; Huang, D. Physicochemical and Functional Properties of Red Lentil Protein Isolates from Three Origins at Different PH. Food Chem. 2021, 358, 129749. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.W.; Mullenix, G.J.; Maynard, C.J.; Lee, J.T.; Rao, S.K.; Butler, L.D.; Orlowski, S.K.; Kidd, M.T. Interactions of the Branched-Chain Amino Acids. 2. Practical Adjustments in Valine and Isoleucine. J. Appl. Poult. Res. 2022, 31, 100241. [Google Scholar] [CrossRef]
- Chetry, N.; Devi, T.G. Intermolecular Interaction Study of L-Threonine in Polar Aprotic Solvent: Experimental and Theoretical Study. J. Mol. Liq. 2021, 338, 116689. [Google Scholar] [CrossRef]
- Haque, E.; Chand, R.; Kapila, S. Biofunctional Properties of Bioactive Peptides of Milk Origin. Food Rev. Int. 2008, 25, 28–43. [Google Scholar] [CrossRef]









| Index | Cheese A * (Control) | Cheese B ** (Test) |
|---|---|---|
| Mass fraction of solids, % | 55.16 ± 0.21 | 54.85 ± 0.23 |
| Mass fraction of fat in solids, % | 48.93 ± 1.95 | 49.50 ± 1.04 |
| Mass fraction of total protein, % | 20.23 ± 0.28 | 21.23 ± 0.31 |
| Mass fraction of salt, % | 2.34 ± 0.72 | 3.13 ± 0.55 |
| pH | 4.74 ± 0.18 | 4.13 ± 0.06 |
| Sample | Fragment in Amino Acid Sequence | Peptide Sequence in one Letter Code * | Bioactivity | Molecular Weight, Da |
|---|---|---|---|---|
| A | 1–5 | MMKSF | 0.730558 | 643.3 |
| A | 1–5 | MKVLI | 0.194789 | 603.4 |
| A | 1–7 | MMSFVSL | 0.488188 | 894.4 |
| A | 7–11 | LVVTI | 0.0498343 | 624.3 |
| A | 8–12 | VVTIL | 0.0759513 | 624.3 |
| A | 11–16 | TCGAQA | 0.162151 | 550.2 |
| A | 16–21 | RPKHPI | 0.396125 | 747.5 |
| B | 20–28 | EQLTKCEVF | 0.154299 | 1176.5 |
| B | 22–31 | KHQGLPQEVL | 0.224327 | 1148.6 |
| A | 22–31 | NVPGEIVESL | 0.118838 | 1056.6 |
| A | 23–28 | TKCEVF | 0.182253 | 806.3 |
| A, B | 31–35 | SQETY | 0.0993374 | 627.3 |
| B | 31–42 | SQETYKQEKNMA | 0.127054 | 1536.6 |
| A | 32–38 | SSSEESI | 0.0916819 | 978.2 |
| A, B | 35–42 | KGYGGVSL | 0.226416 | 780.4 |
| A, B | 38–45 | GGVSLPEW | 0.628953 | 844.4 |
| A, B | 39–43 | FSDKI | 0.403109 | 609.3 |
| A | 39–45 | TRINKKI | 0.0758366 | 872.6 |
| A | 43–50 | INPSKENL | 0.208794 | 914.5 |
| A | 43–50 | PEWVCTTF | 0.703949 | 1142.4 |
| A | 44–50 | NPSKENL | 0.209493 | 801.4 |
| A, B | 46–50 | VCTTF | 0.383259 | 650.2 |
| A | 48–59 | GKEKVNELSKDI | 0.105419 | 1439.7 |
| A, B | 51–62 | CSTFCKEVVRNA | 0.32156 | 1436.6 |
| A | 51–55 | QSAPL | 0.491633 | 515.3 |
| A, B | 51–55 | HTSGY | 0.16794 | 724.2 |
| A, B | 51–59 | HTSGYDTQA | 0.090863 | 1139.3 |
| A | 52–56 | VLSRY | 0.181831 | 637.4 |
| A, B | 54–59 | SRYPSY | 0.442889 | 1012.3 |
| A, B | 55–67 | CKEVVRNANEEEY | 0.0499419 | 1662.7 |
| A | 56–60 | DTQAI | 0.094131 | 547.3 |
| A | 57–61 | PSYGL | 0.748397 | 696.2 |
| B | 60–67 | GSESTEDQAMEDI | 0.0764591 | 1571.5 |
| A, B | 60–68 | GSESTEDQA | 0.0703232 | 1163.3 |
| B | 61–69 | VQNNDSTEY | 0.0623822 | 1149.4 |
| B | 63–69 | NEEEYSI | 0.0502548 | 883.4 |
| A | 64–70 | YQQKPVA | 0.12662 | 913.4 |
| A | 65–70 | QQKPVA | 0.121875 | 670.4 |
| A | 69–73 | QTQSL | 0.105763 | 656.3 |
| B | 70–77 | GSSSEESA | 0.0857066 | 833.3 |
| A | 72–76 | INNQF | 0.436729 | 635.3 |
| A | 73–77 | NNQFL | 0.693175 | 635.3 |
| B | 73–80 | KQMEAESI | 0.0604811 | 1015.4 |
| A | 76–81 | TFPGPIP | 0.774112 | 627.4 |
| A | 78–86 | EVATEEVKI | 0.0379171 | 1017.5 |
| B | 78–86 | ESISSSEEI | 0.0703352 | 1140.4 |
| B | 80–91 | CKDDQNPHSSNI | 0.313549 | 1437.5 |
| B | 80–94 | CKDDQNPHSSNICNI | 0.61108 | 1687.7 |
| B | 87–93 | TVDDKHY | 0.0687443 | 957.4 |
| A | 87–96 | TVDDKHYQKA | 0.0839258 | 1284.6 |
| B | 87–96 | VPNSVEQKHI | 0.119726 | 1150.6 |
| A, B | 87–92 | AVRSPA | 0.171253 | 680.3 |
| A, B | 88–94 | VRSPAQI | 0.149257 | 850.4 |
| A, B | 90–94 | EKTKI | 0.0411874 | 618.4 |
| A, B | 90–96 | EKTKIPA | 0.0778473 | 866.4 |
| A, B | 93–102 | TQTPVVVPPF | 0.213655 | 1164.6 |
| B | 93–103 | TQTPVVVPPFL | 0.336248 | 1197.7 |
| B | 95–100 | SCDKFL | 0.844337 | 712.3 |
| A | 96–100 | QWQVL | 0.584037 | 673.4 |
| A | 100–104 | LDDDL | 0.294973 | 590.3 |
| A | 101–108 | DDDLTDDI | 0.139488 | 921.4 |
| A | 103–109 | LNENKVL | 0.0983577 | 829.5 |
| A | 104–109 | NENKVL | 0.0944044 | 716.4 |
| B | 107–117 | KSCQAQPTTMA | 0.451658 | 1325.5 |
| A | 110–115 | VLDTDY | 0.109603 | 805.3 |
| A, B | 115–119 | RLKKY | 0.162844 | 707.5 |
| A | 116–122 | DKVGINY | 0.13778 | 644.4 |
| A, B | 120–126 | KVPQLEI | 0.109796 | 826.5 |
| A | 122–127 | CMENSA | 0.152971 | 734.2 |
| A, B | 125–131 | DQVKRNA | 0.0817682 | 830.4 |
| A | 127–131 | VPNSA | 0.12024 | 567.2 |
| A | 127–134 | PKYPVEPF | 0.630562 | 976.5 |
| B | 128–133 | EPEQSL | 0.108742 | 702.3 |
| A | 128–134 | EPEQSLA | 0.111383 | 853.3 |
| B | 129–134 | LCSEKL | 0.227722 | 888.4 |
| A, B | 130–134 | CSEKL | 0.263366 | 692.4 |
| A | 130–134 | PVEPF | 0.604782 | 588.3 |
| B | 130–137 | CSEKLDQW | 0.393924 | 579.3 |
| A, B | 135–140 | TESQSL | 0.0615565 | 904.2 |
| A, B | 135–142 | TESQSLTL | 0.0988356 | 1198.3 |
| A | 135–143 | TPTLNREQL | 0.0748829 | 1151.5 |
| B | 136–142 | HSMKEGI | 0.177846 | 801.4 |
| A, B | 138–142 | LCEKL | 0.274007 | 1008.4 |
| A, B | 143–151 | HAQQKEPIM | 0.258844 | 1081.5 |
| A | 148–159 | SGEPTSTPTTEA | 0.0824745 | 1177.5 |
| A | 160–165 | VESTVA | 0.0342661 | 605.3 |
| A, B | 160–167 | VESTVATL | 0.0738447 | 899.4 |
| A, B | 163–176 | TKKTKLTEEEKNRL | 0.0531694 | 922.5 |
| B | 166–172 | SFNPTQL | 0.619614 | 806.4 |
| A | 166–174 | TLEDSPEVI | 0.0723477 | 1162.4 |
| A, B | 167–172 | PPTVMF | 0.826762 | 771.3 |
| A | 168–180 | EDSPEVIESPPEI | 0.166577 | 1440.7 |
| A | 168–172 | NPTQL | 0.293133 | 572.3 |
| A, B | 173–180 | PPQSVLSL | 0.487707 | 840.5 |
| A | 174–178 | YPSGA | 0.433892 | 574.2 |
| A, B | 175–179 | PSGAW | 0.870583 | 597.2 |
| A | 175–180 | ESPPEI | 0.211756 | 671.3 |
| A, B | 179–186 | SLSQSKVL | 0.184348 | 941.5 |
| A | 180–186 | KKISQRY | 0.127085 | 922.5 |
| A, B | 181–186 | SQSKVL | 0.210141 | 741.4 |
| B | 181–189 | TVQVTSTAV | 0.0291752 | 1160.4 |
| A, B | 181–192 | SQPKVLPVPQKA | 0.343103 | 1281.8 |
| A | 182–188 | VPLGTQY | 0.161862 | 937.3 |
| A, B | 183–189 | SQRYQKF | 0.584592 | 1036.5 |
| A, B | 185–191 | GTQYTDA | 0.086917 | 755.3 |
| A | 189–194 | TDAPSF | 0.417727 | 717.2 |
| A | 192–197 | PSFSDI | 0.547239 | 665.3 |
| A, B | 195–201 | SDIPNPI | 0.573163 | 755.4 |
| A | 200–204 | QHQKA | 0.0976463 | 611.3 |
| A, B | 202–213 | IGSENSEKTTMPLW | 0.351255 | 1533.5 |
| A, B | 205–209 | MKPWI | 0.853622 | 674.4 |
| A | 208–213 | YQEPVL | 0.234191 | 748.4 |
| A | 209–216 | IQPKTKVI | 0.108562 | 926.6 |
| B | 210–218 | QPKTKVIPY | 0.213985 | 697.4 |
| A, B | 217–221 | PYVRY | 0.446831 | 1233.6 |
| Sample | Peptide Sequence | Bioactivity * | Potential Bioactive Properties ** | Peptide Structure *** |
|---|---|---|---|---|
| A, B | PSGAW | 0.870583 | ACE inhibitors |  |
| A, B | MKPWI | 0.853622 | ACE inhibitors |  |
| B | SCDKFL | 0.844337 | ACE inhibitor |  |
| A, B | PPTVMF | 0.826762 | DPP-4 inhibitors |  |
| A | TFPGPIP | 0.774112 | DPP-4 inhibitor |  |
| A | PSYGL | 0.748397 | Opioid |  |
| A | MMKSF | 0.730558 | ACE inhibitor |  |
| A | PEWVCTTF | 0.703949 | ACE inhibitor |  |
| A | NNQFL | 0.693175 | DPP-4 inhibitor |  |
| A | PKYPVEPF | 0.630562 | Antioxidant |  |
| A, B | GGVSLPEW | 0.628953 | ACE inhibitors |  |
| B | SFNPTQL | 0.619614 | ACE inhibitor |  |
| B | CKDDQNPHSSNICNI | 0.61108 | Antimicrobial |  |
| A | PVEPF | 0.604782 | Opioid; DPP-4 inhibitor; antioxidant |  |
| A | QWQVL | 0.584037 | Immunomodulating |  |
| A, B | SDIPNPI | 0.573163 | Growth stimulators |  |
| A | PSFSDI | 0.547239 | Anticancer; antimicrobial |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurbanova, M.; Voroshilin, R.; Kozlova, O.; Atuchin, V. Effect of Lactobacteria on Bioactive Peptides and Their Sequence Identification in Mature Cheese. Microorganisms 2022, 10, 2068. https://doi.org/10.3390/microorganisms10102068
Kurbanova M, Voroshilin R, Kozlova O, Atuchin V. Effect of Lactobacteria on Bioactive Peptides and Their Sequence Identification in Mature Cheese. Microorganisms. 2022; 10(10):2068. https://doi.org/10.3390/microorganisms10102068
Chicago/Turabian StyleKurbanova, Marina, Roman Voroshilin, Oksana Kozlova, and Victor Atuchin. 2022. "Effect of Lactobacteria on Bioactive Peptides and Their Sequence Identification in Mature Cheese" Microorganisms 10, no. 10: 2068. https://doi.org/10.3390/microorganisms10102068