A Paradox in Bacterial Pathogenesis: Activation of the Local Macrophage Inflammasome Is Required for Virulence of Streptococcus uberis
Abstract
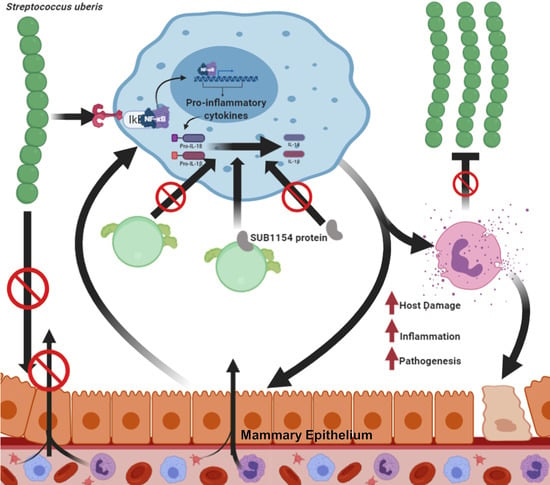
:1. Introduction
2. Results
2.1. Streptococcus uberis Colonisation Requires SUB1154 Protein In Vivo
2.2. Host Cytokine Responses Reflect Overt Clinical Signs
2.3. SUB1154 Elicits Host Antimicrobial Peptide Release and Reduces Epithelial Lining Integrity
2.4. SUB1154 Elicits IL-1β Release from a Novel Ex Vivo Challenge Model in an NLRP3-Dependent Manner
2.5. SUB1154 Alone Does Not Elicit IL-1β from Naïve Macrophage
2.6. The Host Transcriptome Is Reprogrammed in Response to S. uberis Independently of SUB1154
2.7. Downstream Inflammasome Signalling Characterises the SUB1154-Dependent Host Response
3. Discussion
4. Materials and Methods
4.1. Isolation of CD14+ Leukocytes from Bovine Milk
4.2. Bacterial Strains and Culturing Conditions
4.3. Detergent Extraction and Immunoblotting of SUB1154
4.4. Animal Models of Bovine Mastitis
4.5. Ethics Statement
4.6. Intramammary Challenge with S. uberis Strains
4.7. Clinical Scores
4.8. ELISA Analysis of Milk Samples
4.9. Flow Cytometry
4.10. Cytokine Response of Ex Vivo CD14+ Cell Challenged with S. uberis and Recombinant SUB1154 Protein
4.11. Extraction, Purification, and Library Preparation of mRNA from Challenged CD14+ Leukocytes
4.12. Illumina Sequencing and Differential Gene Expression Analysis
4.13. Data Visualisation
4.14. Quantification and Statistical Analysis
4.15. Data and Code Availability
4.16. Materials Availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grace, K.; Harris, C.; Reeves, H.; Healey, K.; Coyne, L.; Borriello, S.P. UK Veterinary Antibiotic Resistance and Sales Surveillance UK-VARSS 2014; Veterinary Medicines Directorate: New Haw, Addlestone, UK, 2014. [Google Scholar]
- Veterinary Medicines Directorate and APHA. UK Veterinary Antibiotic Resistance and Sales Surveillance UK-VARSS 2018; Veterinary Medicines Directorate: New Haw, Addlestone, UK, 2019. [Google Scholar]
- Wilson, D.J.; González, R.N.; Hertl, J.; Schulte, H.F.; Bennett, G.J.; Schukken, Y.H.; Gröhn, Y.T. Effect of clinical mastitis on the lactation curve: A mixed model estimation using daily milk weights. J. Dairy Sci. 2004, 87, 2073–2084. [Google Scholar] [CrossRef]
- Casey, J.W.; Holden, N.M. The Relationship between Greenhouse Gas Emissions and the Intensity of Milk Production in Ireland. J. Environ. Qual. 2005, 34, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Ward, P.N.; Field, T.R.; Jones, C.L.; Lincoln, R.A.; Leigh, J.A. MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect. Immun. 2003, 71, 4842–4849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinarello, C.A. Immunological and Inflammatory Functions of the Interleukin-1 Family. Annu. Rev. Immunol. 2009, 27, 519–550. [Google Scholar] [CrossRef]
- Littlejohn, M.D.; Turner, S.A.; Walker, C.G.; Berry, S.D.; Tiplady, K.; Sherlock, R.G.; Sutherland, G.; Swift, S.; Garrick, D.; Lacy-Hulbert, S.J.; et al. Identification of an immune modulation locus utilising a bovine mammary gland infection challenge model. J. Dairy Res. 2018, 85, 185–192. [Google Scholar] [CrossRef]
- Kostadinova, E.; Chaput, C.; Gutbier, B.; Lippmann, J.; Sander, L.E.; Mitchell, T.J.; Suttorp, N.; Witzenrath, M.; Opitz, B. NLRP3 protects alveolar barrier integrity by an inflammasome-independent increase of epithelial cell adherence. Sci. Rep. 2016, 6, 30943. [Google Scholar] [CrossRef] [Green Version]
- Valderrama, J.A.; Riestra, A.M.; Gao, N.J.; Larock, C.N.; Gupta, N.; Ali, S.R.; Hoffman, H.M.; Ghosh, P.; Nizet, V. Group A streptococcal M protein activates the NLRP3 inflammasome. Nat. Microbiol. 2017, 2, 1425–1434. [Google Scholar] [CrossRef] [Green Version]
- Palazon-Riquelme, P.; Lopez-Castejon, G. The inflammasomes, immune guardians at defence barriers. Immunology 2018, 155, 320–330. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Kuang, M.; Li, L.; Wang, Y.; Yang, F.; Wang, G. UFL1 modulates NLRP3 inflammasome activation and protects against pyroptosis in LPS-stimulated bovine mammary epithelial cells. Mol. Immunol. 2019, 112, 1–9. [Google Scholar] [CrossRef]
- Günther, J.; Czabanska, A.; Bauer, I.; Leigh, J.A.; Holst, O.; Seyfert, H.M.M. Streptococcus uberis strains isolated from the bovine mammary gland evade immune recognition by mammary epithelial cells, but not of macrophages. Vet. Res. 2016, 47, 13. [Google Scholar] [CrossRef] [Green Version]
- Günther, J.; Koy, M.; Berthold, A.; Schuberth, H.J.; Seyfert, H.M. Comparison of the pathogen species-specific immune response in udder derived cell types and their models. Vet. Res. 2016, 47, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyes, K.M.; Drackley, J.K.; Morin, D.E.; Bionaz, M.; Rodriguez-Zas, S.L.; Everts, R.E.; Lewin, H.A.; Loor, J.J. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPAR signaling as potential mechanism for the negative relationships between immune response and lipi. BMC Genom. 2009, 10, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swanson, K.M.; Stelwagen, K.; Dobson, J.; Henderson, H.V.; Davis, S.R.; Farr, V.C.; Singh, K. Transcriptome profiling of Streptococcus uberis-induced mastitis reveals fundamental differences between immune gene expression in the mammary gland and in a primary cell culture model. J. Dairy Sci. 2009, 92, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Schukken, Y.H.H.; Günther, J.; Fitzpatrick, J.; Fontaine, M.C.C.; Goetze, L.; Holst, O.; Leigh, J.; Petzl, W.; Schuberth, H.J.J.; Sipka, A.; et al. Host-response patterns of intramammary infections in dairy cows. Vet. Immunol. Immunopathol. 2011, 144, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Crispi, K.; Atalla, H.; Miglior, F.; Mallard, B.A. Bovine mastitis: Frontiers in immunogenetics. Front. Immunol. 2014, 5, 493. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y. The role of chemokines in neutrophil biology. Front. Biosci. 2008, 13, 2400–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, A.W.; Shears, A.L.; Hibbitt, K.G. The elimination of serum-resistant Escherichia coli from experimentally infected single mammary glands of healthy cows. Res. Vet. Sci. 1978, 25, 89–93. [Google Scholar] [CrossRef]
- Field, T.R.; Ward, P.N.; Pedersen, L.H.; Leigh, J.A. The hyaluronic acid capsule of Streptococcus uberis is not required for the development of infection and clinical mastitis. Infect. Immun. 2003, 71, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Inohara, N.; Ogura, Y.; Fontalba, A.; Gutierrez, O.; Pons, F.; Crespo, J.; Fukase, K.; Inamura, S.; Kusumoto, S.; Hashimoto, M.; et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: Implications for Crohn’s disease. J. Biol. Chem. 2003, 278, 5509–5512. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Xia, P.; Li, S.; Zhang, T.; Wang, T.T.; Zhu, J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 2017, 69, 297–304. [Google Scholar] [CrossRef]
- von Moltke, J.; Ayres, J.S.; Kofoed, E.M.; Chavarría-Smith, J.; Vance, R.E. Recognition of Bacteria by Inflammasomes. Annu. Rev. Immunol. 2013, 31, 73–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Kanneganti, T.D. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J. Cell Biol. 2016, 213, 617–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Guo, H.; Callaway, J.B.; Ting, J.P.Y. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting Edge: NF-κB Activating Pattern Recognition and Cytokine Receptors License NLRP3 Inflammasome Activation by Regulating NLRP3 Expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, V.A.K.; Fitzgerald, K.A. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell 2016, 165, 792–800. [Google Scholar] [CrossRef] [Green Version]
- Soehnlein, O.; Oehmcke, S.; Ma, X.; Rothfuchs, A.G.; Frithiof, R.; Van Rooijen, N.; Mörgelin, M.; Herwald, H.; Lindbom, L. Neutrophil degranulation mediates severe lung damage triggered by streptococcal M1 protein. Eur. Respir. J. 2008, 32, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Egan, S.A.; Kurian, D.; Ward, P.N.; Hunt, L.; Leigh, J.A. Identification of sortase A (SrtA) substrates in Streptococcus uberis: Evidence for an additional hexapeptide (LPXXXD) sorting motif. J. Proteome Res. 2010, 9, 1088–1095. [Google Scholar] [CrossRef] [Green Version]
- Leigh, J.A.A.; Egan, S.A.A.; Ward, P.N.N.; Field, T.R.R.; Coffey, T.J.J. Sortase anchored proteins of Streptococcus uberis play major roles in the pathogenesis of bovine mastitis in dairy cattle. Vet. Res. 2010, 41, 63. [Google Scholar] [CrossRef] [Green Version]
- Ward, P.N.; Holden, M.T.G.; Leigh, J.A.; Lennard, N.; Bignell, A.; Barron, A.; Clark, L.; Quail, M.A.; Woodward, J.; Barrell, B.G.; et al. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genom. 2009, 10, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, D.L.; Ward, P.N.; Rapier, C.D.; Leigh, J.A.; Bowler, L.D. Identification of a differentially expressed oligopeptide binding protein (OppA2) in Streptococcus uberis by representational difference analysis of cDNA. J. Bacteriol. 2003, 185, 5210–5219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deluyker, H.A.; Gay, J.M.; Weaver, L.D. Interrelationships of Somatic Cell Count, Mastitis, and Milk Yield in a Low Somatic Cell Count Herd. J. Dairy Sci. 1993, 76, 3445–3452. [Google Scholar] [CrossRef]
- Bradley, A.J.; Leach, K.A.; Breen, J.E.; Green, L.E.; Green, M.J. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 2007, 160, 253–257. [Google Scholar] [CrossRef] [Green Version]
- Paape, M.J.; Wergin, W.P.; Guidry, A.J.; Schultze, W.D. Phagocytic defense of the ruminant mammary gland. Adv. Exp. Med. Biol. 1981, 137, 555–578. [Google Scholar]
- Leigh, J.A.; Field, T.R. Streptococcus uberis resists the bactericidal action of bovine neutrophils despite the presence of bound immunoglobulin. Infect. Immun. 1994, 62, 1854–1859. [Google Scholar] [CrossRef] [Green Version]
- Lewandowska-Sabat, A.M.; Olsaker, I.; Boman, G.M.; Downing, A.; Talbot, R.; Storset, A.K. The early phase transcriptome of bovine monocyte-derived macrophages infected with Staphylococcus aureus in vitro. BMC Genom. 2013, 14, 891. [Google Scholar] [CrossRef] [Green Version]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef] [Green Version]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Gabay, C.; Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 1999, 340, 448–454. [Google Scholar] [CrossRef]
- Mudaliar, M.; Tassi, R.; Thomas, F.C.; McNeilly, T.N.; Weidt, S.K.; McLaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.D.; et al. Mastitomics, the integrated omics of bovine milk in an experimental model of: Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellnitz, O.; Wall, S.K.; Saudenova, M.; Bruckmaier, R.M. Effect of intramammary administration of prednisolone on the blood-milk barrier during the immune response of the mammary gland to lipopolysaccharide. Am. J. Vet. Res. 2014, 75, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Pisanu, S.; Marogna, G.; Cubeddu, T.; Pagnozzi, D.; Cacciotto, C.; Campesi, F.; Schianchi, G.; Rocca, S.; Uzzau, S. Production and release of antimicrobial and immune defense proteins by mammary epithelial cells following Streptococcus uberis infection of sheep. Infect. Immun. 2013, 81, 3182–3197. [Google Scholar] [CrossRef] [Green Version]
- Cubeddu, T.; Cacciotto, C.; Pisanu, S.; Tedde, V.; Alberti, A.; Pittau, M.; Dore, S.; Cannas, A.; Uzzau, S.; Rocca, S.; et al. Cathelicidin production and release by mammary epithelial cells during infectious mastitis. Vet. Immunol. Immunopathol. 2017, 189, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Coll, R.C.; Robertson, A.A.B.; Chae, J.J.; Higgins, S.C.; Muñoz-Planillo, R.; Inserra, M.C.; Vetter, I.; Dungan, L.S.; Monks, B.G.; Stutz, A.; et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015, 21, 248–257. [Google Scholar] [CrossRef] [Green Version]
- de Araújo, E.F.; Feriotti, C.; de Lima Galdino, N.A.; Preite, N.W.; Calich, V.L.G.; Loures, F.V. The IDO-AhR axis controls Th17/Treg immunity in a pulmonary model of fungal infection. Front. Immunol. 2017, 8, 880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopalakrishna, Y.; Langley, D.; Sarkar, N. Detection of high level of polyadenylate-contining RNA in bacteria by the use of a single-step RNA isoation procedure. Nucleic Acids Res. 1981, 9, 3545–3554. [Google Scholar] [CrossRef] [Green Version]
- Archer, N.; Walsh, M.D.; Shahrezaei, V.; Hebenstreit, D. Modeling Enzyme Processivity Reveals that RNA-Seq Libraries Are Biased in Characteristic and Correctable Ways. Cell Syst. 2016, 3, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lönnerberg, P.; Linnarsson, S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 2014, 11, 163–166. [Google Scholar] [CrossRef]
- Grohmann, U.; Fallarino, F.; Puccetti, P. Tolerance, DCs and tryptophan: Much ado about IDO. Trends Immunol. 2003, 24, 242–248. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Z.; Xiao, T.S. Post-translational regulation of inflammasomes. Cell. Mol. Immunol. 2017, 14, 65–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Calvo, M.; Peterson, E.P.; Leiting, B.; Ruel, R.; Nicholson, D.W.; Thornberry, N.A. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J. Biol. Chem. 1998, 273, 32608–32613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baroja-Mazo, A.; Martín-Sánchez, F.; Gomez, A.I.; Martínez, C.M.; Amores-Iniesta, J.; Compan, V.; Barberà-Cremades, M.; Yagüe, J.; Ruiz-Ortiz, E.; Antón, J.; et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat. Immunol. 2014, 15, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Demirel, I.; Persson, A.; Brauner, A.; Särndahl, E.; Kruse, R.; Persson, K. Activation of the NLRP3 inflammasome pathway by uropathogenic Escherichia coli is virulence factor-dependent and influences colonization of bladder epithelial cells. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, R.G.; Finch, J.M. Phagocytosis of Streptococcus uberis by bovine mammary gland macrophages. Res. Vet. Sci. 1997, 62, 74–78. [Google Scholar] [CrossRef]
- Thomas, L.H.; Haider, W.; Hill, A.W.; Cook, R.S. Pathologic findings of experimentally induced Streptococcus uberis infection in the mammary gland of cows. Am. J. Vet. Res. 1994, 55, 1723–1728. [Google Scholar] [PubMed]
- Taxman, D.J.; Huang, M.T.H.; Ting, J.P.Y. Inflammasome inhibition as a pathogenic stealth mechanism. Cell Host Microbe 2010, 8, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Mathur, A.; Feng, S.; Hayward, J.A.; Ngo, C.; Fox, D.; Atmosukarto, I.I.; Price, J.D.; Schauer, K.; Märtlbauer, E.; Robertson, A.A.B.; et al. A multicomponent toxin from Bacillus cereus incites inflammation and shapes host outcome via the NLRP3 inflammasome. Nat. Microbiol. 2019, 4, 362–374. [Google Scholar] [CrossRef]
- Frodermann, V.; van Duijn, J.; van Puijvelde, G.H.M.; van Santbrink, P.J.; Lagraauw, H.M.; de Vries, M.R.; Quax, P.H.A.; Bot, I.; Foks, A.C.; de Jager, S.C.A.; et al. Heat-killed Staphylococcus aureus reduces atherosclerosis by inducing anti-inflammatory macrophages. J. Intern. Med. 2016, 279, 592–605. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, S.; Reyes-Aldasoro, C.C.; Candel, S.; Renshaw, S.A.; Mulero, V.; Calado, Â. Cxcl8 (IL-8) Mediates Neutrophil Recruitment and Behavior in the Zebrafish Inflammatory Response. J. Immunol. 2013, 190, 4349–4359. [Google Scholar] [CrossRef]
- LaRock, C.N.; Döhrmann, S.; Todd, J.; Corriden, R.; Olson, J.; Johannssen, T.; Lepenies, B.; Gallo, R.L.; Ghosh, P.; Nizet, V. Group A Streptococcal M1 Protein Sequesters Cathelicidin to Evade Innate Immune Killing. Cell Host Microbe 2015, 18, 471–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaRock, D.L.; Russell, R.; Johnson, A.F.; Wilde, S.; LaRock, C.N. Group A Streptococcus Infection of the Nasopharynx Requires Proinflammatory Signaling Through the Interleukin-1 Receptor. Infect. Immun. 2020. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, J.; Rohde, M.; Siemens, N.; Kreikemeyer, B.; Bergman, P.; Johansson, L.; Norrby-Teglund, A. LL-37 Triggers Formation of Streptococcus pyogenes Extracellular Vesicle-Like Structures with Immune Stimulatory Properties. J. Innate Immun. 2016, 8, 243–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, J.N.; Nizet, V. Bacterial Evasion of Host Antimicrobial Peptide Defenses. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, J.A.; Field, T.R.; Williams, M.R. Two strains of Streptococcus uberis, of differing ability to cause clinical mastitis, differ in their ability to resist some host defence factors. Res. Vet. Sci. 1990, 49, 85–87. [Google Scholar] [CrossRef]
- Hill, A.W.; Finch, J.M.; Field, T.R.; Leigh, J.A. Immune modification of the pathogenesis of Streptococcus uberis mastitis in the dairy cow. FEMS Immunol. Med. Microbiol. 1994, 8, 109–117. [Google Scholar] [CrossRef]
- Blanchard, A. The Use of Random Mutagenesis in the Functional Annotation of the Streptococcus Uberis Genome. Ph.D. Thesis, University uf Nottingham, Nottingham, UK, 2015. [Google Scholar]
- An, Z.; Li, J.; Yu, J.; Wang, X.; Gao, H.; Zhang, W.; Wei, Z.; Zhang, J.; Zhang, Y.; Zhao, J.; et al. Neutrophil extracellular traps induced by IL-8 aggravate atherosclerosis via activation NF-κB signaling in macrophages. Cell Cycle 2019, 18, 2928–2938. [Google Scholar] [CrossRef]
- Rivas, A.L.; Tadevosyan, R.; Quimby, F.W.; Coksaygan, T.; Lein, D.H. Identification of subpopulations of bovine mammary-gland phagocytes and evaluation of sensitivity and specificity of morphologic and functional indicators of bovine mastitis. Can. J. Vet. Res. 2002, 66, 165–172. [Google Scholar]
- Addis, M.F.; Bronzo, V.; Puggioni, G.M.G.; Cacciotto, C.; Tedde, V.; Pagnozzi, D.; Locatelli, C.; Casula, A.; Curone, G.; Uzzau, S.; et al. Relationship between milk cathelicidin abundance and microbiologic culture in clinical mastitis. J. Dairy Sci. 2017, 100, 2944–2953. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, P.D.; Kejariwal, A.; Guo, N.; Mi, H.; Campbell, M.J.; Muruganujan, A.; Lazareva-Ulitsky, B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006, 34, W645–W650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilghe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.4.0. BioConductor 2019. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis Using the Grammar of Graphics; Springer: New York, NY, USA, 2016. [Google Scholar]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [Green Version]





| Score | Appearance of Quarter | Score | Appearance of Milk |
|---|---|---|---|
| 1 | Normal | 1 | Normal |
| 2 | Minor changes (e.g., hardness) | 2 | Minor changes (e.g., a few flakes) |
| 3 | Moderate signs (e.g., heat, tenderness) | 3 | Moderate signs (e.g., clots, clumps) |
| 4 | Severe signs (e.g., distended and discomfort on palpation) | 4 | Severe Signs (e.g., changes in colour, composition) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archer, N.; Egan, S.A.; Coffey, T.J.; Emes, R.D.; Addis, M.F.; Ward, P.N.; Blanchard, A.M.; Leigh, J.A. A Paradox in Bacterial Pathogenesis: Activation of the Local Macrophage Inflammasome Is Required for Virulence of Streptococcus uberis. Pathogens 2020, 9, 997. https://doi.org/10.3390/pathogens9120997
Archer N, Egan SA, Coffey TJ, Emes RD, Addis MF, Ward PN, Blanchard AM, Leigh JA. A Paradox in Bacterial Pathogenesis: Activation of the Local Macrophage Inflammasome Is Required for Virulence of Streptococcus uberis. Pathogens. 2020; 9(12):997. https://doi.org/10.3390/pathogens9120997
Chicago/Turabian StyleArcher, Nathan, Sharon A. Egan, Tracey J. Coffey, Richard D. Emes, M. Filippa Addis, Philip N. Ward, Adam M. Blanchard, and James A. Leigh. 2020. "A Paradox in Bacterial Pathogenesis: Activation of the Local Macrophage Inflammasome Is Required for Virulence of Streptococcus uberis" Pathogens 9, no. 12: 997. https://doi.org/10.3390/pathogens9120997