Prevalence, Resistance Patterns and Biofilm Production Ability of Bacterial Uropathogens from Cases of Community-Acquired Urinary Tract Infections in South Italy
Abstract
:1. Introduction
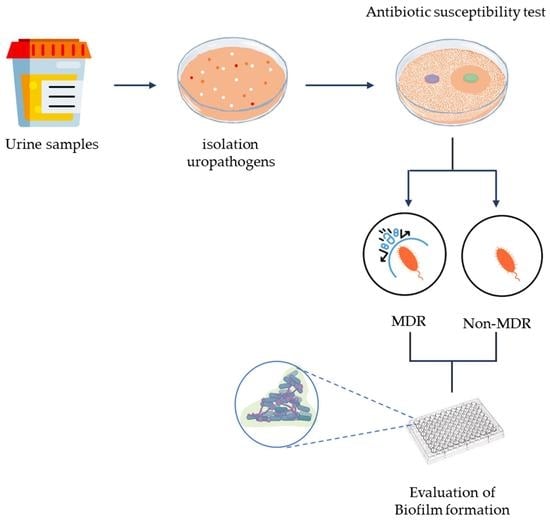
2. Materials and Methods
2.1. Samples Collection
2.2. Inclusion and Exclusion Criteria
2.3. Bacterial Culture, Bacterial Identification, and Antibiotic Susceptibility Test
2.4. Assay of Biofilm Formation
3. Results
3.1. UTI Prevalence in Studied Patients
3.2. Bacteria Implicated in UTI
3.3. Prevalence of Antibiotic Resistance among Identified Uropathogens
3.4. Biofilm Formation of UPs Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, R.D.; Hultgren, S.J. Urinary tract infections: Microbial pathogenesis, host-pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 2020, 18, 211–226. [Google Scholar] [CrossRef]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11, 1756287219832172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kline, K.A.; Lewis, A.L. Gram-Positive Uropathogens, Polymicrobial Urinary Tract Infection, and the Emerging Microbiota of the Urinary Tract. Microbiol. Spectr. 2016, 4, 4.2.04. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomila, A.; Carratalà, J.; Eliakim-Raz, N.; Shaw, E.; Tebé, C.; Wolkewitz, M.; Wiegand, I.; Grier, S.; Vank, C.; Cuperus, N. Clinical outcomes of hospitalised patients with catheter-associated urinary tract infection in countries with a high rate of multidrug-resistance: The COMBACTE-MAGNET RESCUING study. Antimicrob. Resist. Infect. Control 2019, 8, 198. [Google Scholar] [CrossRef] [Green Version]
- Mancini, A.; Pucciarelli, S.; Lombardi, F.E.; Barocci, S.; Pauri, P.; Lodolini, S. Differences between Community—And Hospital—Acquired urinary tract infections in a tertiary care hospital. New Microbiol. 2020, 43, 17–21. [Google Scholar] [PubMed]
- Dell’Annunziata, F.; Ilisso, C.P.; Dell’Aversana, C.; Greco, G.; Coppola, A.; Martora, F.; Dal Piaz, F.; Donadio, G.; Falanga, A.; Galdiero, M.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Influence the miRNA Expression Profile in Human Bronchial Epithelial BEAS-2B Cells. Microorganisms 2020, 8, 1985. [Google Scholar] [CrossRef] [PubMed]
- Storme, O.; Tirán Saucedo, J.; Garcia-Mora, A.; Dehesa-Dávila, M.; Naber, K.G. Risk factors and predisposing conditions for urinary tract infection. Ther. Adv. Urol. 2019, 11, 1756287218814382. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, Y.; Kim, H.; Kim, J.; Koo, S.H.; Kwon, G.C. Rapid Screening of Urinary Tract Infection and Discrimination of Gram-Positive and Gram-Negative Bacteria by Automated Flow Cytometric Analysis Using Sysmex UF-5000. J. Clin. Microbiol. 2018, 56, e02004-17. [Google Scholar] [CrossRef] [Green Version]
- Bitew, A.; Molalign, T.; Chanie, M. Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC Infect. Dis. 2017, 17, 654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi-Te, C.; Shigemura, K.; Nishimoto, K.; Yamada, N.; Kitagawa, K.; Sung, S.Y.; Chen, K.C.; Fujisawa, M. Urinary tract infection pathogens and antimicrobial susceptibilities in Kobe, Japan and Taipei, Taiwan: An international analysis. J. Int. Med. Res. 2020, 48, 300060519867826. [Google Scholar] [CrossRef] [PubMed]
- Dell’Annunziata, F.; Folliero, V.; Giugliano, R.; De Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.R.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef]
- Beahm, N.P.; Nicolle, L.E.; Bursey, A.; Smyth, D.J.; Tsuyuki, R.T. The assessment and management of urinary tract infections in adults: Guidelines for pharmacists. Can. Pharm. J. 2017, 150, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Dune, T.J.; Price, T.K.; Hilt, E.E.; Thomas-White, K.J.; Kliethermes, S.; Brincat, C.; Brubaker, L.; Schreckenberger, P.; Wolfe, A.J.; Mueller, E.R. Urinary Symptoms and Their Associations with Urinary Tract Infections in Urogynecologic Patients. Obstet. Gynecol. 2017, 130, 718–725. [Google Scholar] [CrossRef]
- Dryden, S.D.; Anastasova, S.; Satta, G.; Thompson, A.J.; Leff, D.R.; Darzi, A. Rapid uropathogen identification using surface enhanced Raman spectroscopy active filters. Sci. Rep. 2021, 11, 8802. [Google Scholar] [CrossRef] [PubMed]
- Akoachere, J.F.; Yvonne, S.; Akum, N.H.; Seraphine, E.N. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res. Notes 2012, 5, 219. [Google Scholar] [CrossRef] [Green Version]
- Donkor, E.S.; Horlortu, P.Z.; Dayie, N.T.; Obeng-Nkrumah, N.; Labi, A.K. Community acquired urinary tract infections among adults in Accra, Ghana. Infect. Drug Resist. 2019, 12, 2059–2067. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Petrillo, F.; Pignataro, D.; Di Lella, F.M.; Reibaldi, M.; Fallico, M.; Castellino, N.; Parisi, G.; Trotta, M.C.; D’Amico, M.; Santella, B.; et al. Antimicrobial Susceptibility Patterns and Resistance Trends of Staphylococcus aureus and Coagulase-Negative Staphylococci Strains Isolated from Ocular Infections. Antibiotics 2021, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, F.; Folliero, V.; Santella, B.; Franci, G.; Foglia, F.; Trotta, M.C.; Della Rocca, M.T.; Avitabile, T.; Gagliano, C.; Galdiero, M. Prevalence and Antibiotic Resistance Patterns of Ocular Bacterial Strains Isolated from Pediatric Patients in University Hospital of Campania “Luigi Vanvitelli,” Naples, Italy. Int. J. Microbiol. 2020, 2020, 8847812. [Google Scholar] [CrossRef]
- Holm, A.; Cordoba, G.; Aabenhus, R. Prescription of antibiotics for urinary tract infection in general practice in Denmark. Scand. J. Prim. Health Care 2019, 37, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleidorn, J.; Hummers-Pradier, E.; Schmiemann, G.; Wiese, B.; Gágyor, I. Recurrent urinary tract infections and complications after symptomatic versus antibiotic treatment: Follow-up of a randomised controlled trial. Ger. Med. Sci. 2016, 14, Doc01. [Google Scholar] [CrossRef]
- White, W.L. Erratum to: Why I hate the index finger. Hand 2011, 6, 233. [Google Scholar] [CrossRef] [Green Version]
- López Romo, A.; Quirós, R. Appropriate use of antibiotics: An unmet need. Ther. Adv. Urol. 2019, 11, 1756287219832174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, K.; Hooton, T.M.; Naber, K.G.; Wullt, B.; Colgan, R.; Miller, L.G.; Moran, G.J.; Nicolle, L.E.; Raz, R.; Schaeffer, A.J.; et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 2011, 52, e103–e120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; EUCAST: Växjö, Sweden, 2019. [Google Scholar]
- Davarzani, F.; Saidi, N.; Besharati, S.; Saderi, H.; Rasooli, I.; Owlia, P. Evaluation of Antibiotic Resistance Pattern, Alginate and Biofilm Production in Clinical Isolates of Pseudomonas aeruginosa. Iran. J. Public Health 2021, 50, 341–349. [Google Scholar] [CrossRef]
- de Alteriis, E.; Maselli, V.; Falanga, A.; Galdiero, S.; Di Lella, F.M.; Gesuele, R.; Guida, M.; Galdiero, E. Efficiency of gold nanoparticles coated with the antimicrobial peptide indolicidin against biofilm formation and development of Candida spp. clinical isolates. Infect. Drug Resist. 2018, 11, 915–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Bischoff, S.; Walter, T.; Gerigk, M.; Ebert, M.; Vogelmann, R. Empiric antibiotic therapy in urinary tract infection in patients with risk factors for antibiotic resistance in a German emergency department. BMC Infect. Dis. 2018, 18, 56. [Google Scholar] [CrossRef]
- Petca, R.C.; Negoiță, S.; Mareș, C.; Petca, A.; Popescu, R.I.; Chibelean, C.B. Heterogeneity of Antibiotics Multidrug-Resistance Profile of Uropathogens in Romanian Population. Antibiotics 2021, 10, 523. [Google Scholar] [CrossRef]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Cortes-Penfield, N.W.; Trautner, B.W.; Jump, R.L.P. Urinary Tract Infection and Asymptomatic Bacteriuria in Older Adults. Infect. Dis. Clin. N. Am. 2017, 31, 673–688. [Google Scholar] [CrossRef] [Green Version]
- Martora, F.; Pinto, F.; Folliero, V.; Cammarota, M.; Dell’Annunziata, F.; Squillaci, G.; Galdiero, M.; Morana, A.; Schiraldi, C.; Giovane, A.; et al. Isolation, characterization and analysis of pro-inflammatory potential of Klebsiella pneumoniae outer membrane vesicles. Microb. Pathog. 2019, 136, 103719. [Google Scholar] [CrossRef] [PubMed]
- Cantisani, M.; Vitiello, M.; Falanga, A.; Finamore, E.; Galdiero, M.; Galdiero, S. Peptides complementary to the active loop of porin P2 from Haemophilus influenzae modulate its activity. Int. J. Nanomed. 2012, 7, 2361. [Google Scholar]
- Tandan, M.; Duane, S.; Cormican, M.; Murphy, A.W.; Vellinga, A. Reconsultation and Antimicrobial Treatment of Urinary Tract Infection in Male and Female Patients in General Practice. Antibiotics 2016, 5, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folliero, V.; Caputo, P.; Della Rocca, M.T.; Chianese, A.; Galdiero, M.; Iovene, M.R.; Hay, C.; Franci, G.; Galdiero, M. Prevalence and Antimicrobial Susceptibility Patterns of Bacterial Pathogens in Urinary Tract Infections in University Hospital of Campania “Luigi Vanvitelli” between 2017 and 2018. Antibiotics 2020, 9, 215. [Google Scholar] [CrossRef]
- Groopman, J.D.; Donahue, K.F. Aflatoxin, a human carcinogen: Determination in foods and biological samples by monoclonal antibody affinity chromatography. Assoc. Off. Anal. Chem. 1988, 71, 861–867. [Google Scholar] [CrossRef]
- Pironti, C.; Dell’Annunziata, F.; Giugliano, R.; Folliero, V.; Galdiero, M.; Ricciardi, M.; Motta, O.; Proto, A.; Franci, G. Comparative analysis of peracetic acid (PAA) and permaleic acid (PMA) in disinfection processes. Sci. Total Environ. 2021, 797, 149206. [Google Scholar] [CrossRef]
- Lee, D.S.; Lee, S.J.; Choe, H.S. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. Biomed Res. Int. 2018, 2018, 7656752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odoki, M.; Almustapha Aliero, A.; Tibyangye, J.; Nyabayo Maniga, J.; Wampande, E.; Drago Kato, C.; Agwu, E.; Bazira, J. Prevalence of Bacterial Urinary Tract Infections and Associated Factors among Patients Attending Hospitals in Bushenyi District, Uganda. Int. J. Microbiol. 2019, 2019, 4246780. [Google Scholar] [CrossRef]
- Paris, D.; Caltagirone, M.; Minzulli, P.; Valzano, A.; Ferrara, F.E.O.; Magliano, E. Epidemiology and antibiotic resistance in community-acquired lower urinary tract infections in the Milan area. Microbiol. Med. 2020, 35, 26–31. [Google Scholar] [CrossRef]
- Erdem, I.; Ali, R.K.; Ardic, E.; Omar, S.E.; Mutlu, R.; Topkaya, A.E. Community-acquired lower urinary tract infections: Etiology, antimicrobial resistance, and treatment results in female patients. J. Glob. Infect. Dis. 2018, 10, 129. [Google Scholar] [CrossRef]
- de Oliveira, W.D.; Barboza, M.G.L.; Faustino, G.; Inagaki, W.T.Y.; Sanches, M.S.; Kobayashi, R.K.T.; Vespero, E.C.; Rocha, S.P.D. Virulence, resistance and clonality of Proteus mirabilis isolated from patients with community-acquired urinary tract infection (CA-UTI) in Brazil. Microb. Pathog. 2021, 152, 104642. [Google Scholar] [CrossRef] [PubMed]
- Sarnelli Bruno, D.A.F.; Giuseppe, S.; Ugo, T.; Antonella, G. Report on Antibiotic Resistance and Use of Antibiotic of Antibiotics Detected in the Hospital Structures of Campania. Available online: http://www.regione.campania.it/regione/it/tematiche/antibiotico-resistenza-ed-infezioni-correlate-all-assistenza-64in (accessed on 2 January 2023).
- González, M.J.; Zunino, P.; Scavone, P.; Robino, L. Selection of effective antibiotics for uropathogenic Escherichia coli intracellular bacteria reduction. Front. Cell. Infect. Microbiol. 2020, 10, 542755. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.J.; Barreira, J.C.M.; Carvalho, I.; Trinta, L.; Perreira, L.; Ferreira, I.; Pintado, M. Propensity for biofilm formation by clinical isolates from urinary tract infections: Developing a multifactorial predictive model to improve antibiotherapy. J. Med. Microbiol. 2014, 63, 471–477. [Google Scholar] [CrossRef]
- Ahmad, S. Multiple Drug Resistance Pattern in Urinary Tract Infection Patients in Saudi Arabia. Bangladesh J. Infect. Dis. 2019, 6, 3–7. [Google Scholar] [CrossRef]
- Behzadi, P.; Zsoldiné Urbán, E.; Gajdács, M. Association between Biofilm-Production and Antibiotic Resistance in Uropathogenic Escherichia coli (UPEC). Diseases 2020, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Godbole, G.P.; Cerruto, N.; Chavada, R. Principles of assessment and management of urinary tract infections in older adults. J. Pharm. Pract. Res. 2020, 50, 276–283. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, H.; Bi, D.; Khaledi, A.; Qiao, M. A systematic review and meta-analysis of antibiotic resistance patterns, and the correlation between biofilm formation with virulence factors in uropathogenic E. coli isolated from urinary tract infections. Microb. Pathog. 2020, 144, 104196. [Google Scholar] [CrossRef] [PubMed]
- Meade, E.; Savage, M.; Garvey, M. Effective antimicrobial solutions for eradicating multi-resistant and β-lactamase-producing nosocomial gram-negative pathogens. Antibiotics 2021, 10, 1283. [Google Scholar] [CrossRef]
- Fusco, A.; Coretti, L.; Savio, V.; Buommino, E.; Lembo, F.; Donnarumma, G. Biofilm Formation and Immunomodulatory Activity of Proteus mirabilis Clinically Isolated Strains. Int. J. Mol. Sci. 2017, 18, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raya, S.; Belbase, A.; Dhakal, L.; Govinda Prajapati, K.; Baidya, R.; Kishor Bimali, N. In-Vitro Biofilm Formation and Antimicrobial Resistance of Escherichia coli in Diabetic and Nondiabetic Patients. BioMed Res. Int. 2019, 2019, 1474578. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, C.J., Jr.; Mende, K.; Beckius, M.L.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 2013, 13, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arafa, S.H.; Alshehri, W.A.; Organji, S.R.; Elbanna, K.; Obaid, N.A.; Aldosari, M.S.; Asiri, F.H.; Ahmad, I.; Abulreesh, H.H. Antimicrobial Resistance, Virulence Factor-Encoding Genes, and Biofilm-Forming Ability of Community-Associated Uropathogenic in Western Saudi Arabia. Pol. J. Microbiol. 2022, 71, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Tewawong, N.; Kowaboot, S.; Pimainog, Y.; Watanagul, N.; Thongmee, T.; Poovorawan, Y. Distribution of phylogenetic groups, adhesin genes, biofilm formation, and antimicrobial resistance of uropathogenic Escherichia coli isolated from hospitalized patients in Thailand. PeerJ 2020, 8, e10453. [Google Scholar] [CrossRef] [PubMed]
| Character | n (%) | ||
|---|---|---|---|
| No pathogenic bacteria | 1702 (62.1) | ||
| Pathogenic bacteria | 1039 (37.9) | ||
| Gram + | 39 (3.8) | ||
| Gram − | 1000 (96.2) | ||
| Gender | n (%) | ||
| Female | 760 (73.1) | ||
| Male | 279 (26.9) | ||
| Age Groups n (%) | |||
| Male | Female | Tot. | Age |
| 37 (13.3) | 35 (4.6) | 73 (7.0) | <1 |
| 10 (7.3) | 18 (2.4) | 28 (2.7) | 2–5 |
| 12 (4.3) | 22 (2.9) | 34 (3.3) | 6–12 |
| 19 (6.8) | 42 (5.5) | 61 (5.9) | 13–18 |
| 46 (16.1) | 97 (12.8) | 142 (13.3) | 19–45 |
| 23 (8.2) | 182 (24.0) | 205 (19.7) | 46–60 |
| 184 (66.0) | 312 (41.0) | 4 96 (47.7) | >61 |
| Species | Gram Classification | Number | Prevalence (%) |
|---|---|---|---|
| E. coli | negative | 750 | 72.2 |
| K. pneumoniae | negative | 128 | 12.3 |
| P. mirabilis | negative | 93 | 8.9 |
| E. fecalis | negative | 14 | 1.3 |
| P. aeruginosa | negative | 12 | 1.2 |
| C. koseri | negative | 8 | 0.7 |
| R. plancticola | negative | 1 | 0.1 |
| E. durans | positive | 1 | 0.1 |
| L. amnigena | positive | 1 | 0.1 |
| S. alactolycus | positive | 1 | 0.1 |
| S. agalactiae | positive | 1 | 0.1 |
| S. aureus | positive | 5 | 0.5 |
| E. gallinarum | positive | 5 | 0.5 |
| E. faecium | positive | 5 | 0.5 |
| S. pyogenes | positive | 3 | 0.3 |
| E. aerogenes | negative | 1 | 0.1 |
| S. marcescens | negative | 2 | 0.2 |
| K. oxytoca | negative | 2 | 0.2 |
| S. xylosus | positive | 2 | 0.2 |
| R. ornithiolytica | negative | 1 | 0.1 |
| S. saprophyticus | positive | 1 | 0.1 |
| A. baumannii | negative | 1 | 0.1 |
| S. haemolyticus | positive | 1 | 0.1 |
| Bacteria | Antimicrobial Agents Tested n (%) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amp | Nor | Amc | Cip | Sxt | Ctx | Caz | Gm | Fep | P/T | Fos | Nit | Ert | Imi | Ak | Mem | |
| E. coli | 485 | 358 | 305 | 264 | 231 | 173 | 143 | 104 | 79 | 55 | 49 | 23 | 6 | 1 | 1 | 1 |
| (64.7) | (47.7) | (40.7) | (35.2) | (30.8) | (23.2) | (19.1) | (13.9) | (10.5) | (7.3) | (6.5) | (3.1) | (0.8) | (0.1) | (0.1) | (0.1) | |
| K. pneumoniae | 128 | 52 | 54 | 42 | 44 | 31 | 22 | 9 | 15 | 20 | 31 | - | 3 | 2 | 2 | 2 |
| (100) | (40.6) | (42.2) | (32.8) | (34.4) | (24.2) | (17.1) | (7) | (11.7) | (15.6) | (24.2) | (2.3) | (1.6) | (1.6) | (1.6) | ||
| P. mirabilis | 62 | - | 35 | 36 | 38 | 21 | 16 | 24 | 9 | 8 | 27 | 24 | 7 | 8 | 4 | 2 |
| (66.7) | (37.6) | (38.7) | (40.9) | (22.6) | (17.2) | (26.1) | (9.7) | (8.6) | (29) | (25.8) | (7.5) | (8.6) | (4.3) | (2.2) | ||
| Total (971) | 675 | 410 | 394 | 342 | 313 | 225 | 195 | 137 | 103 | 83 | 107 | 46 | 16 | 11 | 7 | 5 |
| (69.5) | (46.7) | (40.6) | (35.2) | (32.2) | (23.2) | (20.1) | (14.1) | (10.6) | (8.5) | (11.0) | (4.7) | (1.7) | (1.1) | (0.7) | (0.5) | |
| Variable | MDR (33) | Non-MDR (27) | p Value |
|---|---|---|---|
| Biofilm | |||
| Biofilm production | 33 (100%) | 17 (62.5%) | <0.0001 |
| Strong | 15 (45.4%) | 1 (5.8%) | <0.0001 |
| Moderate | 14 (42.4%) | 7 (25.9%) | ns |
| Weak | 4 (12.1%) | 9 (33.3%) | 0.0319 |
| Negative | 0 (0%) | 10 (37.0%) | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maione, A.; Galdiero, E.; Cirillo, L.; Gambino, E.; Gallo, M.A.; Sasso, F.P.; Petrillo, A.; Guida, M.; Galdiero, M. Prevalence, Resistance Patterns and Biofilm Production Ability of Bacterial Uropathogens from Cases of Community-Acquired Urinary Tract Infections in South Italy. Pathogens 2023, 12, 537. https://doi.org/10.3390/pathogens12040537
Maione A, Galdiero E, Cirillo L, Gambino E, Gallo MA, Sasso FP, Petrillo A, Guida M, Galdiero M. Prevalence, Resistance Patterns and Biofilm Production Ability of Bacterial Uropathogens from Cases of Community-Acquired Urinary Tract Infections in South Italy. Pathogens. 2023; 12(4):537. https://doi.org/10.3390/pathogens12040537
Chicago/Turabian StyleMaione, Angela, Emilia Galdiero, Luigi Cirillo, Edvige Gambino, Maria Assunta Gallo, Francesca Paola Sasso, Arianna Petrillo, Marco Guida, and Marilena Galdiero. 2023. "Prevalence, Resistance Patterns and Biofilm Production Ability of Bacterial Uropathogens from Cases of Community-Acquired Urinary Tract Infections in South Italy" Pathogens 12, no. 4: 537. https://doi.org/10.3390/pathogens12040537