Assembly and Analysis of Haemonchus contortus Transcriptome as a Tool for the Knowledge of Ivermectin Resistance Mechanisms
Abstract
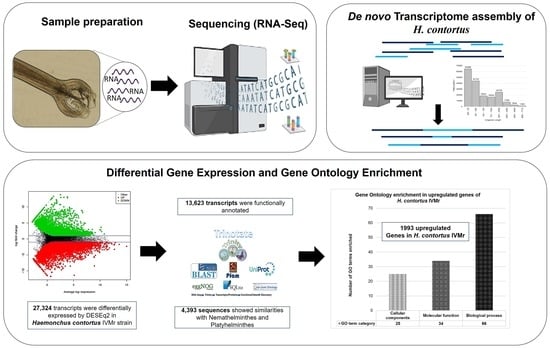
:1. Introduction
2. Materials and Methods
2.1. History of Haemonchus contortus Isolates
Parasites
2.2. RNA-Seq
2.2.1. Preparation of Haemonchus contortus Total RNA for Transcriptome Sequencing
2.2.2. Preparation of cDNA Libraries for the Sequencing Platform
2.2.3. Assembly, Annotation and Differential Expression Analysis
2.2.4. Gene Ontology Enrichment Analysis (GOEA)
3. Results and Discussion
3.1. Transcriptome Assembly and Features
3.2. Functional Transcription Annotation of H. contortus
3.3. Differential Gene Expression between Two Mexican H. contortus Strains
3.4. Gene Ontology Enrichment Analysis (GOEA)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez-Vivas, R.I.; Grisi, L.; Pérez, L.A.A.; Silva, V.H.; Torres-Acosta, F.J.; Fragoso, S.H.; Romero, S.D.; Rosario, C.R.; Saldierna, F.; García, C.D. Potential economic impact assessment for cattle parasites in Mexico. Review. Rev. Mex. Cienc. Pecu. 2017, 8, 61–74. [Google Scholar] [CrossRef]
- Craig, T.M. Gastrointestinal nematodes, diagnosis and control. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 185–199. [Google Scholar] [CrossRef]
- Abosse, J.S.; Terefe, G.; Teshale, B.M. Comparative study on pathological changes in sheep and goats experimentally infected with Haemonchus contortus. Surg. Exp. Pathol. 2022, 5, 14. [Google Scholar] [CrossRef]
- Miller, C.M.; Waghorn, T.S.; Leathwick, D.M.; Candy, P.M.; Oliver, A.M.; Watson, T.G. The production cost of anthelmintic resistance in lambs. Vet. Parasitol. 2012, 186, 376–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Tamayo, A.A.; López-Arellano, M.E.; González-Garduño, R.; Torres-Hernández, G.; De la Mora-Valle, A.; Becerril-Pérez, C.; Hernández-Mendo, O.; Ramírez-Bribiesca, E.; Huchin-Cab, M. Haemonchus contortus infection induces a variable immune response in resistant and susceptible Pelibuey sheep. Vet. Immunol. Immunopathol. 2021, 234, 110218. [Google Scholar] [CrossRef]
- El-Ashram, S.; Al Nasr, I.; Mehmood, R.; Hu, M.; He, L.; Suo, X. Haemonchus contortus and Ovine Host: A Retrospective Review. Int. J. Adv. Res. 2017, 5, 972–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotze, A.C.; Prichard, K. Anthelmintic resistance in Haemonchus contortus: History, mechanisms and diagnosis. In Haemonchus contortus and Haemonchosis—Past, Present and Future Trends; Gasser, R.B., Von Samson-Himmelstjerna, G., Eds.; Elsevier Ltd.: London, UK, 2016; pp. 397–428. [Google Scholar] [CrossRef]
- Flay, K.J.; Hill, F.I.; Muguiro, D.H. A Review: Haemonchus contortus Infection in Pasture-Based Sheep Production Systems, with a Focus on the Pathogenesis of Anaemia and Changes in Haematological Parameters. Animals 2022, 12, 1238. [Google Scholar] [CrossRef]
- Baltrušis, P.; Doyle, S.R.; Halvarsson, P.; Höglund, J. Genome-wide analysis of the response to ivermectin treatment by a Swedish field population of Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2022, 18, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Ardelli, B.F. Transport proteins of the ABC systems superfamily and their role in drug action and resistance in nematodes. Parasitol. Int. 2013, 62, 639–646. [Google Scholar] [CrossRef]
- Kellerová, P.; Matoušková, P.; Lamka, J.; Vokřál, I.; Szotáková, B.; Zajíčková, M.; Pasák, M.; Skálová, L. Ivermectin-induced changes in the expression of cytochromes P450 and efflux transporters in Haemonchus contortus female and male adults. Vet. Parasitol. 2019, 273, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.J.; Robertson, A.P.; Choudhary, S. Ivermectin: An anthelmintic, an insecticide, and much more. Trends Parasitol. 2021, 37, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Redman, E.; Packard, E.; Grillo, V.; Smith, J.; Jackson, F.; Gilleard, J.S. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. Int. J. Parasitol. 2008, 38, 111–122. [Google Scholar] [CrossRef]
- Whittaker, J.H.; Carlson, S.A.; Jones, D.E.; Brewer, M.T. Molecular mechanisms for anthelmintic resistance in strongyle nematode parasites of veterinary importance. J. Vet. Pharmacol. Ther. 2017, 40, 105–115. [Google Scholar] [CrossRef]
- Laing, R.; Martinelli, A.; Tracey, A.; Holroyd, N.; Gilleard, J.S.; Cotton, J.A. Haemonchus contortus: Genome Structure, Organization and Comparative Genomics. Adv Parasitol. 2016, 93, 569–598. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.R.; Tracey, A.; Laing, R.; Holroyd, N.; Bartley, D.; Bazant, W.; Beasley, H.; Beech, R.; Britton, C.; Brooks, K.; et al. Genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus, a model gastrointestinal worm. Comms. Biol. 2020, 3, 656. [Google Scholar] [CrossRef]
- Reyes-Guerrero, D.E.; Cedillo-Borda, M.; Alonso-Morales, R.A.; Alonso-Diaz, M.A.; Olmedo-Juarez, A.; Mendoza-de-Gives, P.; Lopez-Arellano, M.E. Comparative study of transcription profiles of the P-glycoprotein transporters of two Haemonchus contortus isolates: Susceptible and resistant to ivermectin. Mol. Biochem. Parasitol. 2020, 238, 111281. [Google Scholar] [CrossRef] [PubMed]
- Sallé, G.; Doyle, S.R.; Cortet, J.; Cabaret, J.; Berriman, M.; Holroyd, N.; Cotton, J.A. The global diversity of Haemonchus contortus is shaped by human intervention and climate. Nat. Comm. 2019, 10, 4811. [Google Scholar] [CrossRef] [Green Version]
- Kebeta, M.M.; Hine, B.C.; Walkden-Brown, S.W.; Kahn, L.P.; Piedrafita, D.P.; Bailey, S.; Doyle, E.K. Investigation of the combined efficacy of two Haemonchus contortus vaccines in weaner Merino sheep. Vet. Parasitol. 2022, 301, 109637. [Google Scholar] [CrossRef]
- Campos-Ruelas, R.; Herrera-Rodríguez, D.; Quiroz-Romero, H.; Olazarán-Jenkins, S. Resistencia de Haemonchus contortus a bencimidazoles en ovinos de México. Tec. Pecu. Mex. 1990, 28, 30–34. [Google Scholar]
- González-Garduño, R.; Torres-Hernández, G.; López-Arellano, M.E.; Mendoza-de-Gives, P. Resistencia antihelmíntica de nematodos parásitos en ovinos. Rev. Geogra. Agrícol. 2012, 48–49, 63–74. [Google Scholar]
- Schwarz, E.M.; Korhonen, P.K.; Campbell, B.E.; Young, N.D.; Jex, A.R.; Jabbar, A.; Hall, R.S.; Mondal, A.; Howe, A.C.; Pell, J.; et al. The genome and developmental transcriptome of the Strongylid nematode Haemonchus contortus. Genome Biol. 2013, 14, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mate, L.; Ballent, M.; Canton, C.; Lanusse, C.; Ceballos, L.; Alvarez L, L.I.; Liron, J. ABC-transporter gene expression in ivermectin-susceptible and resistant Haemonchus contortus isolates. Vet. Parasitol. 2022, 302, 109647. [Google Scholar] [CrossRef] [PubMed]
- Cedillo-Borda, M.; López-Arellano, M.E.; Reyes-Guerrero, D.E. In vitro assessment of ivermectin resistance and gene expression profiles of P-glycoprotein genes from Haemonchus contortus (L3). Bio-101 2020, 10, e3851. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 10 December 2019).
- Krueger, F.; James, F.; Ewels, P.; Afyounian, E.; Schuster-Boeckler, B. FelixKrueger/TrimGalore; v0.6.7—DOI via Zenodo (0.6.7); Zenodo: Geneva, Switzerland, 2021. [Google Scholar] [CrossRef]
- Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Laing, R.; Kikuchi, T.; Martinelli, A.; Tsai, I.J.; Beech, R.N.; Redman, E.; Holroyd, N.; Bartley, D.J.; Beasley, H.; Britton, C.; et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013, 14, R88. [Google Scholar] [CrossRef] [Green Version]
- Palevich, N.; Maclean, P.H.; Baten, A.; Scott, R.W.; Leathwick, D.M. The Genome Sequence of the Anthelmintic- Susceptible New Zealand Haemonchus contortus. Genome Biol. Evol. 2019, 11, 1965–1970. [Google Scholar] [CrossRef] [Green Version]
- Ponstingl, H.; Ning, Z. SMALT—A new mapper for DNA sequencing reads. F1000Posters 2010, 1, 313. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Jacinto, V.; Sanchez-Flores, A.; Vega-Alvarado, L. Integrative Differential Expression Analysis for Multiple EXperiments (IDEAMEX): A Web Server Tool for Integrated RNA-Seq Data Analysis. Front. Genet. 2019, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezansoff, A.M.; Laing, R.; Martinelli, A.; Stasiuk, S.; Redman, E.; Bartley, D.; Holroyd, N.; Devaney, E.; Sargison, N.D.; Doyle, S.; et al. The confounding effects of high genetic diversity on the determination and interpretation of differential gene expression analysis in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2019, 49, 847–858. [Google Scholar] [CrossRef]
- Cantacessi, C.; Campbell, B.E.; Young, N.D.; Jex, A.R.; Hall, R.S.; Presidente, P.J.A.; Zawadzki, J.; Zhong, W.; Aleman-Meza, B.; Loukas, A.; et al. Differences in transcription between free-living and CO2-activated third-stage larvae of Haemonchus contortus. BMC Genom. 2010, 11, 266. Available online: http://www.biomedcentral.com/1471-2164/11/266 (accessed on 13 April 2021). [CrossRef] [PubMed] [Green Version]
- Emery, D.L.; Hunt, P.W.; Le Jambre, L.F. Haemonchus contortus: The then and now, and where to from here? Int. J. Parasitol. 2016, 46, 755–769. [Google Scholar] [CrossRef] [Green Version]
- Stasiuk, S.J.; MacNevin, G.; Workentine, M.L.; Gray, D.; Redman, E.; Bartley, D.; Morrison, A.; Sharma, N.; Colwell, D.; Ro, D.K.; et al. Similarities and differences in the biotransformation and transcriptomic responses of Caenorhabditis elegans and Haemonchus contortus to five different benzimidazole drugs. Int. J. Parasitol. Drugs Drug Resist. 2019, 11, 13–29. [Google Scholar] [CrossRef]
- Skuce, P.J.; Yaga, R.; Lainson, F.A.; Knox, D.P. An evaluation of serial analysis of gene expression (SAGE) in the parasitic nematode, Haemonchus contortus. Parasitology 2005, 130, 553–559. [Google Scholar] [CrossRef]
- El-Abdellati, A.; De Graef, J.; Van Zeveren, A.; Donnan, A.; Skuce, P.; Walsh, T.; Wolstenholme, A.; Tait, A.; Vercruysse, J.; Claerebout, E.; et al. Altered avr-14B gene transcription patterns in ivermectin-resistant isolates of the cattle parasites, Cooperia oncophora and Ostertagia ostertagi. Int. J. Parasitol. 2011, 41, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.M.; Storey, B.; Howell, S.; Harper, K.M.; Kaplan, R.M.; Wolstenholme, A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011, 180, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Beech, R.; Levitt, N.; Cambos, M.; Zhou, S.; Forrester, S. Association of ion-channel genotype and macrocyclic lactone sensitivity traits in Haemonchus contortus. Mol. Biochem. Parasitol. 2010, 171, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.P.; Hayashi, J.; Beech, R.N.; Prichard, R.K. Study of the nematode putative GABA type-A receptor subunits: Evidence for modulation by ivermectin. J. Neurochem. 2002, 83, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.; Che, H.; Beech, R.N.; Prichard, R.K. Characterization of Haemonchus contortus P-glycoprotein-16 and its interaction with the macrocyclic lactone anthelmintics. Mol. Biochem. Parasitol. 2015, 204, 11–15. [Google Scholar] [CrossRef]
- Bonilla-Suárez, H.A.; Olazarán-Jenkins, S.; Reyes-Guerrero, D.E.; Maza-Lopez, J.; Olmedo-Juárez, A.; Mendoza-de-Gives, P.; López-Arellano, M.E. P-glycoprotein gene expression analysis of ivermectin resistance in sheep naturally infected with Haemonchus contortus. Mex. J. Biotechnol. 2022, 7, 16–31. [Google Scholar] [CrossRef]
- Laing, R.; Doyle, S.R.; McIntyre, J.; Maitland, K.; Morrison, A.; Bartley, D.J.; Kaplan, R.; Chaudhry, U.; Sargison, N.; Tait, A.; et al. Transcriptomic analyses implicate neuronal plasticity and chloride homeostasis in ivermectin resistance and response to treatment in a parasitic nematode. PLoS Pathog. 2022, 18, e1010545. [Google Scholar] [CrossRef]
- Jex, A.R.; Gasser, R.B.; Schwarz, E.M. Transcriptomic Resources for Parasitic Nematodes of Veterinary Importance. Trends Parasitol. 2019, 35, 72–84. [Google Scholar] [CrossRef]
- Roeber, F.; Jex, A.R.; Gasser, R.B. Next-generation molecular-diagnostic tools for gastrointestinal nematodes of livestock, with an emphasis on small ruminants: A turning point? Adv. Parasitol. 2013, 83, 267–333. [Google Scholar] [CrossRef]
- Lu, M.R.; Lai, C.K.; Liao, B.Y.; Tsai, I.J. Comparative Transcriptomics across Nematode Life Cycles Reveal Gene Expression Conservation and Correlated Evolution in Adjacent Developmental Stages. Genome Biol. Evol. 2020, 12, 1019–1030. [Google Scholar] [CrossRef]
- Glendinning, S.K.; Buckingham, S.D.; Sattelle, D.B.; Wonnacott, S.; Wolstenholme, A.J. Glutamate-gated chloride channels of Haemonchus contortus restore drug sensitivity to ivermectin resistant Caenorhabditis elegans. PLoS ONE 2011, 6, e22390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, S.M.; Wolstenholme, A.J. P-glycoproteins of Haemonchus contortus: Development of real-time PCR assays for gene expression studies. J. Helminthol. 2012, 86, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Abubucker, S.; Zarlenga, D.S.; Martin, J.; Yin, Y.; Wang, Z.; McCarter, J.P.; Gasbarre, L.; Wilson, R.K.; Mitreva, M. The transcriptomes of the cattle parasitic nematode Ostertagia ostartagi. Vet. Parasitol. 2009, 162, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Zhang, Y.; Wu, S.; Wang, Z.; Tuersong, W.; Wang, C.; Liu, F.; Hu, M. Genome-Wide Identification of CircRNAs of Infective Larvae and Adult Worms of Parasitic Nematode, Haemonchus contortus. Front. Cell. Infect. Microbiol. 2021, 11, 764089. [Google Scholar] [CrossRef]
- Tuersong, W.; Zhou, C.; Wu, S.; Qin, P.; Wang, C.; Di, W.; Liu, L.; Liu, H.; Hu, M. Comparative analysis on transcriptomics of ivermectin resistant and susceptible strains of Haemonchus contortus. Parasit. Vectors 2022, 15, 159. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 8, D325–D334. [Google Scholar] [CrossRef]
- Issouf, M.; Guégnard, F.; Koch, C.; Le Vern, Y.; Blanchard-Letort, A.; Che, H.; Beech, R.N.; Kerboeuf, D.; Neveu, C. Haemonchus contortus P-glycoproteins interact with host eosinophil granules: A novel insight into the role of ABC transporters in host-parasite interaction. PLoS ONE 2014, 9, e87802. [Google Scholar] [CrossRef] [Green Version]
- Ménez, C.; Alberich, M.; Courtot, E.; Guegnard, F.; Blanchard, A.; Aguilaniu, H.; Lespine, A. The transcription factor NHR-8: A new target to increase ivermectin efficacy in nematodes. PLoS Pathog. 2019, 15, e1007598. [Google Scholar] [CrossRef] [Green Version]
- Maizels, R.M.; Gomez-Escobar, N.; Gregory, W.F.; Murray, J.; Zang, X. Immune evasion genes from filarial nematodes. Int. J. Parasitol. 2001, 31, 889–898. [Google Scholar] [CrossRef] [Green Version]



| Features and Statistics | |
|---|---|
| Status | High-quality transcriptome |
| Isolation source | Male adult nematode (ovine abomasum) |
| Assembly method | Trinity/RNA-Seq de novo assembly |
| Transcriptome coverage | 150× |
| Sequencing technology | Illumina |
| Assembled transcriptome size (bp) | 126,974,821 (~127 Mb) |
| Total number of unigen transcripts | 77,422 |
| All transcripts contigs N50 (bp) | 1149 |
| DNA G + C (%) content | 44.4 |
| DNA A + T (%) content | 55.6 |
| Total of Enriched Gene Ontology Terms | LFC ≤ −1 and ≥ 1 (p ≤ 0.05) | LFC ≤ −2 and ≥ 2 (p ≤ 0.01) | ||
|---|---|---|---|---|
| Total of Upregulated Genes | ||||
| IVMr | IVMs | IVMr | IVMs | |
| 1993 | 1929 | 1241 | 835 | |
| Molecular Function | 34 | 33 | 29 | 22 |
| Biological Process | 66 | 31 | 50 | 28 |
| Cellular Component | 25 | 29 | 17 | 10 |
| GO Term | Gene | Trinity ID Transcript | Fold Change | p Value | Definition Based on Gene Ontology Consortium [59,60] |
|---|---|---|---|---|---|
| Molecular Function terms | |||||
| GO:0015562 Efflux transmembrane transporter activity | PGP1 | DN7767 | 7.60 | 3.97 × 10−16 | Enables the transfer of a specific substance or related group of substances from inside of the cell to outside of the cell across a membrane |
| PGP3 | DN40711 | 2.62 | 0.00035 | ||
| GO:0008559 ABC-type xenobiotic transporter activity | PGP1 | DN7767 | 7.60 | 3.97 × 10−16 | Catalysis of the reaction: ATP + H2O + xenobiotic (in) = ADP + phosphate + xenobiotic (out) |
| PGP3 | DN40711 | 2.62 | 0.00035 | ||
| GO:0042626 ATPase-coupled transmembrane transporter activity | PGP1 | DN7767 | 7.60 | 3.97 × 10−16 | Primary active transporter of a solute across a membrane, via the reaction: ATP + H2O = ADP + phosphate, to directly drive the transport of a substance across a membrane. The transport protein may be transiently phosphorylated (P-type transporters), or not (ABC-type transporters) |
| CED7 | DN133 | 2.07 | 1.84 × 10−7 | ||
| WHT1 | DN5402 | 3.22 | 2.38 × 10−16 | ||
| ABCB8 | DN8534 | 1.32 | 0.00021 | ||
| MRP2 | DN14918 | 1.81 | 0.00046 | ||
| ABCA4 | DN48845 | 2.09 | 0.00262 | ||
| ABCD2 | DN5820 | 2.96 | 1.85 × 10−9 | ||
| Biological Process terms | |||||
| GO:0093002 Response to nematicide activity | PGP1 | DN7767 | 7.60 | 3.97 × 10−16 | Processes that result in a change in state or activity of a cell or an organism (movement, secretion, enzyme production, gene expression, etc.) as a result of a nematicide stimulus |
| SEK1 | DN2949 | 2.03 | 3.24 × 10−10 | ||
| KGB1 | DN7108 | 2.33 | 1.39 × 10−12 | ||
| PMK1 | DN7240 | 2.03 | 1.62 × 10−10 | ||
| NSY1 | DN4471 | 1.36 | 0.00035 | ||
| GO:0043050 Pharyngeal nematode pumping | NAS5 | DN1871 | 3.55 | 4.97 × 10−27 | Contraction and relaxation movements of the pharyngeal muscle that mediate feeding in nematodes |
| PXL1 | DN10500 | 9.06 | 6.63 × 10−12 | ||
| MYO4 | DN4583 | 1.29 | 0.00962 | ||
| ATPB | DN2532 | 1.62 | 4.50 × 10−7 | ||
| CSK1 | DN2039 | 1.35 | 3.02 × 10−6 | ||
| GO:0045887 Positive regulation of synaptic assembly | BLI4 | DN5118 | 2.69 | 3.99 × 10−45 | Processes that activate or increase the frequency, rate or extent of synaptic assembly at neuromuscular junction |
| CBPE | DN6090 | 1.32 | 6.43 × 10−7 | ||
| UN104 | DN30477 | 1.74 | 0.00083 | ||
| NEC2 | DN8717 | 1.15 | 0.00152 | ||
| GO:0008152 metabolic process | NHR-8 | DN3547 | 1.15 | 6.12 × 10−7 | Chemical reactions and pathways by which living organisms transform chemical substances. Additionally, they include macromolecular processes (DNA repair and replication, protein synthesis and degradation) |
| YMX8 | DN1007 | 2.91 | 1.19 × 10−7 | ||
| Cellular Components terms | |||||
| GO:0043231 Intracellular membrane-bounded organelle | NHR-8 | DN3547 | 1.15 | 6.12 × 10−7 | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell |
| GO:0016020 Membrane | PGP1 | DN10250 | 9.94 | 1.29 × 10−15 | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it |
| WHT3 | DN10777 | 2.42 | 2.76 × 10−10 | ||
| GO Term | Gene | Trinity ID Transcript | Fold Change | p Value | Definition Based on Gene Ontology Consortium [59,60] |
|---|---|---|---|---|---|
| Molecular Function terms | |||||
| GO:0003777 Microtubule motor activity | HUM6 | DN10330 | 3.25 | 1.94 × 10−6 | Motor activity that generates movement along a microtubule, driven by ATP hydrolysis |
| KLC | DN11269 | 8.36 | 1.94 × 10−9 | ||
| KI26L | DN12477 | 1.71 | 1.98 × 10−5 | ||
| KLP3 | DN15949 | 1.34 | 0.00012 | ||
| MYO1 | DN19625 | 3.36 | 7.09 × 10−6 | ||
| MYO3 | DN32812 | 2.88 | 2.34 × 10−17 | ||
| MYO4 | DN4583 | 1.29 | 0.00962 | ||
| Biological Process terms | |||||
| GO:0014722 Regulation of skeletal muscle contraction | UNC89 | DN2825 | 2.28 | 5.90 × 10−7 | Processes that modulate the frequency, rate or extent of skeletal muscle contraction by changing the calcium ion signals that trigger contraction |
| GO:0007018 Microtubule-based movement | HUM6 | DN10330 | 3.25 | 1.94 × 10−6 | Microtubule-based processes that result in the movement of organelles, other microtubules, or other cellular components by polymerization or depolymerization of microtubules |
| KLC | DN11269 | 8.36 | 1.94 × 10−9 | ||
| KI26L | DN12477 | 1.71 | 1.98 × 10−5 | ||
| KLP3 | DN15949 | 1.34 | 0.00012 | ||
| MYO1 | DN19625 | 3.36 | 7.09 × 10−6 | ||
| MYO3 | DN32812 | 2.88 | 2.34 × 10−17 | ||
| MYO4 | DN4583 | 1.29 | 0.00962 | ||
| GO:0040017 Positive regulation of locomotion | UNC89 | DN2825 | 2.28 | 5.90 × 10−7 | Any process that activates or increases the frequency, rate or extent of locomotion of a cell or organism |
| UNC80 | DN13535 | 1.59 | 0.00046 | ||
| UNC52 | DN11715 | 1.24 | 0.00893 | ||
| LRP | DN12558 | 3.02 | 0.00744 | ||
| EPI1 | DN15848 | 1.55 | 0.00384 | ||
| UNC6 | DN19598 | 2.03 | 0.00016 | ||
| UNC22 | DN2116 | 1.68 | 1.41 × 10−6 | ||
| Cellular Components terms | |||||
| GO:0016021 Integral component of membrane | PGP1 | DN10250 | 9.94 | 1.29 × 10−15 | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it |
| NPL21 | DN10287 | 1.61 | 0.00057 | ||
| VEM1 | DN1029 | 2.04 | 2.12 × 10−12 | ||
| SKAT1 | DN1033 | 1.59 | 3.53 × 10−10 | ||
| NHX9 | DN1038 | 1.71 | 4.57 × 10−9 | ||
| PGAP2 | DN10392 | 3.09 | 5.80 × 10−22 | ||
| GO Term | Gene | Trinity ID Transcript | Fold Change | p Value | Definition Based on Gene Ontology Consortium [59,60] |
|---|---|---|---|---|---|
| Molecular Function terms | |||||
| GO:0045735 nutrient reservoir activity GO:0005319 lipid transporter activity | VIT1 | DN59905 | 4.87 | 0.00024 | GO:0045735 Functions in the storage of nutritious substrates. GO:0005319 Enables the directed movement of lipids into, out of or within a cell, or between cells |
| VIT2 | DN60842 | 12.87 | 1.52 × 10−9 | ||
| VIT4 | DN68591 | 13.68 | 2.60 × 10−59 | ||
| VIT5 | DN53584 | 5.27 | 0.00042 | ||
| VIT6 | DN61483 | 8.20 | 1.90 × 10−9 | ||
| GO:0004129 cytochrome-c oxidase activity | COX1 | DN43209 | 3.59 | 2.49 × 10−8 | Catalysis of the reaction: 4 ferrocytochrome c + O2 + 4 H+ = 4 ferricytochrome c + 2H2O |
| COX2 | DN37556 | 7.87 | 3.64 × 10−157 | ||
| COX3 | DN59338 | 2.15 | 7.95 × 10−8 | ||
| COX5A | DN6162 | 1.17 | 7.68 × 10−16 | ||
| COX6A | DN3177 | 1.06 | 9.47 × 10−17 | ||
| GO:0055077 gap junction hemi-channel activity | INX3 | DN7528 | 3.90 | 0.00054 | A wide pore channel activity that enables the transport of solutes across the membrane via a gap junction hemi-channel. Two gap junction hemi-channels coupled together form a complete gap junction. |
| INX7 | DN11374 | 2.16 | 2.65 × 10−12 | ||
| INX10 | DN3762 | 1.19 | 2.21 × 10−5 | ||
| INX14 | DN20 | 1.01 | 0.00226 | ||
| Biological Process terms | |||||
| GO:0048557 embryonic digestive tract morphogenesis | LIN53 | DN1263 | 2.25 | 2.62 × 10−14 | The process in which the anatomical structures of the digestive tract are generated and organized during embryonic development. |
| LIN35 | DN4658 | 2.32 | 6.71 × 10−7 | ||
| MIG5 | DN5547 | 2.30 | 1.18× 10−9 | ||
| GO:0040025 vulval development | SQV7 | DN2801 | 2.09 | 6.36 × 10−9 | The process whose specific outcome is the progression of the egg-laying organ of female and hermaphrodite nematodes over time, from its formation to the mature structure. |
| UNC84 | DN2062 | 1.52 | 6.31 × 10−7 | ||
| GLD3 | DN1099 | 1.05 | 0.00104 | ||
| GO:0006458 de novo protein folding | TCPA | DN16687 | 1.39 | 1.92 × 10−10 | The process of assisting in the folding of a nascent peptide chain into its correct tertiary structure. |
| TCPB | DN7949 | 1.66 | 1.45 × 10−7 | ||
| TCPH | DN2283 | 1.28 | 6.37 × 10−6 | ||
| GO:0090727 positive regulation of brood size | CDK4 | DN12924 | 2.28 | 3.38 × 10−10 | Any process that increases brood size. Brood size is the number of progenies that survive embryogenesis and are cared for at one time. |
| CED9 | DN1092 | 2.81 | 1.47 × 10−9 | ||
| CCND | DN1960 | 2.46 | 3.47 × 10−8 | ||
| Cellular Components terms | |||||
| GO:0000932 P-body | PATR1 | DN2981 | 1.37 | 4.87 × 10−8 | A focus on the cytoplasm where mRNAs may become inactivated by decapping or some other mechanism. Protein and RNA localized to these foci are involved in mRNA degradation, nonsense-mediated mRNA decay (NMD), translational repression and RNA-mediated gene silencing. |
| CNOT7 | DN6681 | 1.28 | 2.45 × 10−6 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes-Guerrero, D.E.; Jiménez-Jacinto, V.; Alonso-Morales, R.A.; Alonso-Díaz, M.Á.; Maza-Lopez, J.; Camas-Pereyra, R.; Olmedo-Juárez, A.; Higuera-Piedrahita, R.I.; López-Arellano, M.E. Assembly and Analysis of Haemonchus contortus Transcriptome as a Tool for the Knowledge of Ivermectin Resistance Mechanisms. Pathogens 2023, 12, 499. https://doi.org/10.3390/pathogens12030499
Reyes-Guerrero DE, Jiménez-Jacinto V, Alonso-Morales RA, Alonso-Díaz MÁ, Maza-Lopez J, Camas-Pereyra R, Olmedo-Juárez A, Higuera-Piedrahita RI, López-Arellano ME. Assembly and Analysis of Haemonchus contortus Transcriptome as a Tool for the Knowledge of Ivermectin Resistance Mechanisms. Pathogens. 2023; 12(3):499. https://doi.org/10.3390/pathogens12030499
Chicago/Turabian StyleReyes-Guerrero, David Emanuel, Verónica Jiménez-Jacinto, Rogelio Alejandro Alonso-Morales, Miguel Ángel Alonso-Díaz, Jocelyn Maza-Lopez, René Camas-Pereyra, Agustín Olmedo-Juárez, Rosa Isabel Higuera-Piedrahita, and María Eugenia López-Arellano. 2023. "Assembly and Analysis of Haemonchus contortus Transcriptome as a Tool for the Knowledge of Ivermectin Resistance Mechanisms" Pathogens 12, no. 3: 499. https://doi.org/10.3390/pathogens12030499