Resistance in the Genus Spodoptera: Key Insect Detoxification Genes
Abstract
:Simple Summary
Abstract
1. Introduction
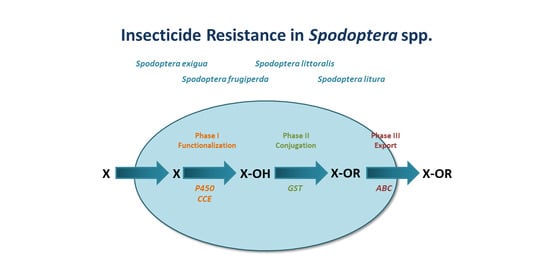
2. Phase I: Functionalization
2.1. Cytochrome P450s
2.1.1. Phylogeny of CYP9A
2.1.2. Resistance through Over-Expression of CYP9A
2.2. Carboxyl/Cholinesterases (CCEs)
2.2.1. Resistance through Over-Expression of CCE
2.2.2. Metabolic Resistance through Point Mutations of CCEs
2.2.3. Target Site Resistance through Point Mutations; the Case of Acetylcholinesterases
3. Phase II: Conjugation
3.1. Glutathione-S-Transferases (GSTs)
3.1.1. Phylogeny of GST Epsilon
3.1.2. GST Activity against Insecticides
3.1.3. Resistance through Over-Expression of GST Epsilon
3.1.4. Resistance through Point Mutations in GST Epsilon
4. Phase III: Elimination/Export
4.1. ATP-Binding Cassette Transporters (ABCs)
4.1.1. Resistance through Point Mutations in ABCs, the Case of ABCC2
4.1.2. Resistance through Over- or Reduced Expression of ABCs
4.1.3. Resistance and Regulation of ABCs Expression
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheng, T.; Wu, J.; Wu, Y.; Chilukuri, R.V.; Huang, L.; Yamamoto, K.; Feng, L.; Li, W.; Chen, Z.; Guo, H.; et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 2017. [Google Scholar] [CrossRef]
- Stokstad, E. New crop pest takes Africa at lightning speed. Science 2017, 356, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.J. Migration and life history strategy of the fall armyworm Spodoptera frugiperda in the Western hemisphere. Insect Sci. Appl. 1987, 8, 543–549. [Google Scholar] [CrossRef]
- Westbrook, J.K. Noctuid migration in Texas within the nocturnal aeroecological boundary layer. Integr. Comp. Biol. 2008, 48, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparks, T.C.; Crossthwaite, A.J.; Nauen, R.; Banba, S.; Cordova, D.; Earley, F.; Ebbinghaus-Kintscher, U.; Fujioka, S.; Hirao, A.; Karmon, D.; et al. Insecticides, biologics and nematicides: Updates to IRAC’s mode of action classification-a tool for resistance management. Pestic. Biochem. Physiol. 2020, 167, 104587. [Google Scholar] [CrossRef]
- Riveron, J.M.; Yunta, C.; Ibrahim, S.S.; Djouaka, R.; Irving, H.; Menze, B.D.; Ismail, H.M.; Hemingway, J.; Ranson, H.; Albert, A.; et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014, 15, R27. [Google Scholar] [CrossRef] [Green Version]
- Feyereisen, R.; Dermauw, W.; van Leeuwen, T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 2015, 121, 61–77. [Google Scholar] [CrossRef]
- Schmidt, J.M.; Good, R.T.; Appleton, B.; Sherrard, J.; Raymant, G.C.; Bogwitz, M.R.; Martin, J.; Daborn, P.J.; Goddard, M.E.; Batterham, P.; et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010, 6, e1000998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devonshire, A.L.; Field, L.M. Gene amplification and insecticide resistance. Annu. Rev. Entomol. 1991, 36, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Field, L.M.; Blackman, R.L.; Tyler-Smith, C.; Devonshire, A.L. Relationship between amount of esterase and gene copy number in insecticide-resistant Myzus persicae (Sulzer). Biochem. J. 1999, 339 Pt 3, 737–742. [Google Scholar] [CrossRef]
- Hu, B.; Huang, H.; Hu, S.; Ren, M.; Wei, Q.; Tian, X.; Elzaki, M.E.A.; Bass, C.; Su, J.; Palli, S.R. Changes in both trans- and cis-regulatory elements mediate insecticide resistance in a lepidopteron pest, Spodoptera exigua. PLoS Genet. 2021, 17, e1009403. [Google Scholar] [CrossRef] [PubMed]
- Amezian, D.; Nauen, R.; le Goff, G. Transcriptional regulation of xenobiotic detoxification genes in insects-An overview. Pestic. Biochem. Physiol. 2021, 174, 104822. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, REVIEWS3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansuy, D. The great diversity of reactions catalyzed by cytochromes P450. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998, 121, 5–14. [Google Scholar] [CrossRef]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochem. Soc. Trans. 2006, 34 Pt 6, 1256–1260. [Google Scholar] [CrossRef]
- Qiu, Y.; Tittiger, C.; Wicker-Thomas, C.; le Goff, G.; Young, S.; Wajnberg, E.; Fricaux, T.; Taquet, N.; Blomquist, G.J.; Feyereisen, R. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 14858–14863. [Google Scholar] [CrossRef] [Green Version]
- Kirkness, E.F.; Haas, B.J.; Sun, W.; Braig, H.R.; Perotti, M.A.; Clark, J.M.; Lee, S.H.; Robertson, H.M.; Kennedy, R.C.; Elhaik, E.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Liu, N. Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS ONE 2011, 6, e29418. [Google Scholar] [CrossRef] [Green Version]
- Nebert, D.W.; Adesnik, M.; Coon, M.J.; Estabrook, R.W.; Gonzalez, F.J.; Guengerich, F.P.; Gunsalus, I.C.; Johnson, E.F.; Kemper, B.; Levin, W.; et al. The P450 gene superfamily: Recommended nomenclature. DNA 1987, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Nelson, D.R.; Coon, M.J.; Estabrook, R.W.; Feyereisen, R.; Fujii-Kuriyama, Y.; Gonzalez, F.J.; Guengerich, F.P.; Gunsalus, I.C.; Johnson, E.F.; et al. The P450 superfamily: Update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991, 10, 1–14. [Google Scholar] [CrossRef]
- Nelson, D.R. Metazoan cytochrome P450 evolution. Comp. Biochem Physiol C Pharmacol Toxicol Endocrinol 1998, 121, 15–22. [Google Scholar] [CrossRef]
- Dermauw, W.; van Leeuwen, T.; Feyereisen, R. Diversity and evolution of the P450 family in arthropods. Insect Biochem. Mol. Biol. 2020, 127, 103490. [Google Scholar] [CrossRef]
- Le Goff, G.; Boundy, S.; Daborn, P.J.; Yen, J.L.; Sofer, L.; Lind, R.; Sabourault, C.; Madi-Ravazzi, L.; ffrench-Constant, R.H. Microarray analysis of cytochrome P450 mediated insecticide resistance in Drosophila. Insect Biochem. Mol. Biol. 2003, 33, 701–708. [Google Scholar] [CrossRef]
- Daborn, P.J.; Lumb, C.; Boey, A.; Wong, W.; Ffrench-Constant, R.H.; Batterham, P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem. Mol. Biol. 2007, 37, 512–519. [Google Scholar] [CrossRef] [PubMed]
- D’Alencon, E.; Sezutsu, H.; Legeai, F.; Permal, E.; Bernard-Samain, S.; Gimenez, S.; Gagneur, C.; Cousserans, F.; Shimomura, M.; Brun-Barale, A.; et al. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc. Natl. Acad. Sci. USA 2010, 107, 7680–7685. [Google Scholar] [CrossRef] [Green Version]
- Feyereisen, R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta 2011, 1814, 19–28. [Google Scholar] [CrossRef]
- Gimenez, S.; Abdelgaffar, H.; le Goff, G.; Hilliou, F.; Blanco, C.A.; Hanniger, S.; Bretaudeau, A.; Legeai, F.; Negre, N.; Jurat-Fuentes, J.L.; et al. Adaptation by copy number variation increases insecticide resistance in the fall armyworm. Commun. Biol. 2020, 3, 664. [Google Scholar] [CrossRef] [PubMed]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagne, N.; Darboux, I.; et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef] [PubMed]
- Kergoat, G.J.; Goldstein, P.Z.; le Ru, B.; Meagher, R.L., Jr.; Zilli, A.; Mitchell, A.; Clamens, A.L.; Gimenez, S.; Barbut, J.; Negre, N.; et al. A novel reference dated phylogeny for the genus Spodoptera Guenee (Lepidoptera: Noctuidae: Noctuinae): New insights into the evolution of a pest-rich genus. Mol. Phylogenet. Evol. 2021, 107161. [Google Scholar] [CrossRef]
- Klai, K.; Chenais, B.; Zidi, M.; Djebbi, S.; Caruso, A.; Denis, F.; Confais, J.; Badawi, M.; Casse, N.; Khemakhem, M.M. Screening of Helicoverpa armigera mobilome revealed transposable element insertions in insecticide resistance genes. Insects 2020, 11, 879. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Tang, W.; He, W.; Ma, X.; Vasseur, L.; Baxter, S.W.; Yang, G.; Huang, S.; Song, F.; You, M. Characterization and expression of the cytochrome P450 gene family in diamondback moth, Plutella xylostella (L.). Sci. Rep. 2015, 5, 8952. [Google Scholar] [CrossRef] [PubMed]
- Carareto, C.M.; Hernandez, E.H.; Vieira, C. Genomic regions harboring insecticide resistance-associated Cyp genes are enriched by transposable element fragments carrying putative transcription factor binding sites in two sibling Drosophila species. Gene 2014, 537, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilding, C.S.; Smith, I.; Lynd, A.; Yawson, A.E.; Weetman, D.; Paine, M.J.; Donnelly, M.J. A cis-regulatory sequence driving metabolic insecticide resistance in mosquitoes: Functional characterisation and signatures of selection. Insect Biochem. Mol. Biol. 2012, 42, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Guo, Z.; Riegler, M.; Xi, Z.; Liang, G.; Xu, Y. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 2017, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Giraudo, M.; Hilliou, F.; Fricaux, T.; Audant, P.; Feyereisen, R.; le Goff, G. Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): Responses to plant allelochemicals and pesticides. Insect Mol. Biol. 2015, 24, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, H.; Liu, Z.; Wu, S.; Yang, Y.; Feyereisen, R.; Heckel, D.G.; Wu, Y. Phylogenetic and functional characterization of ten P450 genes from the CYP6AE subfamily of Helicoverpa armigera involved in xenobiotic metabolism. Insect Biochem. Mol. Biol. 2018, 93, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.L.; Goh, D.; Thompson, D.M.; Verma, K.D.; Heckel, D.G.; Gahan, L.J.; Roe, R.M.; Hodgson, E. Cytochrome P450 (CYP)9A1 in Heliothis virescens: The first member of a new CYP family. Insect Biochem. Mol. Biol. 1997, 27, 605–615. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, S.; Wu, S.; Yue, L.; Wu, Y. Constitutive overexpression of multiple cytochrome P450 genes associated with pyrethroid resistance in Helicoverpa armigera. J. Econ. Entomol. 2006, 99, 1784–1789. [Google Scholar] [CrossRef]
- Yang, Y.; Yue, L.; Chen, S.; Wu, Y. Functional expression of Helicoverpa armigera CYP9A12 and CYP9A14 in Saccharomyces cerevisiae. Pestic. Biochem. Physiol. 2008, 92, 101–105. [Google Scholar] [CrossRef]
- Boaventura, D.; Buer, B.; Hamaekers, N.; Maiwald, F.; Nauen, R. Toxicological and molecular profiling of insecticide resistance in a Brazilian strain of fall armyworm resistant to Bt Cry1 proteins. Pest. Manag. Sci. 2020. [Google Scholar] [CrossRef]
- Do Nascimento, A.R.; Fresia, P.; Consoli, F.L.; Omoto, C. Comparative transcriptome analysis of lufenuron-resistant and susceptible strains of Spodoptera frugiperda (Lepidoptera: Noctuidae). BMC Genom. 2015, 16, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafeez, M.; Liu, S.; Yousaf, H.K.; Jan, S.; Wang, R.L.; Fernandez-Grandon, G.M.; Li, X.; Gulzar, A.; Ali, B.; Rehman, M.; et al. RNA interference-mediated knockdown of a cytochrome P450 gene enhanced the toxicity of alpha-cypermethrin in xanthotoxin-fed larvae of Spodoptera exigua (Hubner). Pestic. Biochem. Physiol. 2020, 162, 6–14. [Google Scholar] [CrossRef]
- Hafeez, M.; Liu, S.; Jan, S.; Shi, L.; Fernandez-Grandon, G.M.; Gulzar, A.; Ali, B.; Rehman, M.; Wang, M. Knock-down of gossypol-inducing cytochrome P450 genes reduced deltamethrin sensitivity in Spodoptera exigua (Hubner). Int. J. Mol. Sci. 2019, 20, 2248. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.L.; Liu, S.W.; Baerson, S.R.; Qin, Z.; Ma, Z.H.; Su, Y.J.; Zhang, J.E. Identification and functional analysis of a novel cytochrome P450 gene CYP9A105 associated with pyrethroid detoxification in Spodoptera exigua Hubner. Int. J. Mol. Sci. 2018, 19, 737. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Mei, Y.; Liu, R.; Chen, X.; Li, D.; Wang, C. Transcriptome analysis of Spodoptera litura reveals the molecular mechanism to pyrethroids resistance. Pestic. Biochem. Physiol. 2020, 169, 104649. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Staehelin, C.; Xia, Q.Q.; Su, Y.J.; Zeng, R.S. Identification and characterization of CYP9A40 from the tobacco cutworm moth (Spodoptera litura), a cytochrome P450 gene induced by plant allelochemicals and insecticides. Int. J. Mol. Sci. 2015, 16, 22606–22620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakeshott, J.; Claudianos, C.; Campbell, P.M.; Newcomb, R.; Russell, R.J. Biochemical genetics and genomics of insect esterases. In Comprehensive Molecular Insect Science-Pharmacology; Gilbert, L.I., Iatrou, K., Gill, S.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 309–381. [Google Scholar]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Oakeshott, J.G.; Devonshire, A.L.; Claudianos, C.; Sutherland, T.D.; Horne, I.; Campbell, P.M.; Ollis, D.L.; Russell, R.J. Comparing the organophosphorus and carbamate insecticide resistance mutations in cholin- and carboxyl-esterases. Chem. Biol. Interact. 2005, 157–158, 269–275. [Google Scholar] [CrossRef]
- Oakeshott, J.G.; Johnson, R.M.; Berenbaum, M.R.; Ranson, H.; Cristino, A.S.; Claudianos, C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol. Biol. 2010, 19 (Suppl. 1), 147–163. [Google Scholar] [CrossRef] [Green Version]
- Pearce, S.L.; Clarke, D.F.; East, P.D.; Elfekih, S.; Gordon, K.H.J.; Jermiin, L.S.; McGaughran, A.; Oakeshott, J.G.; Papanikolaou, A.; Perera, O.P.; et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017, 15, 63. [Google Scholar] [CrossRef] [Green Version]
- Durand, N.; Carot-Sans, G.; Chertemps, T.; Montagne, N.; Jacquin-Joly, E.; Debernard, S.; Maibeche-Coisne, M. A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol. Biol. 2010, 19, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.; Chertemps, T.; Maibeche-Coisne, M. Antennal carboxylesterases in a moth, structural and functional diversity. Commun. Integr. Biol. 2012, 5, 284–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlin, C.; Rosell, G.; Carot-Sans, G.; Francois, M.C.; Bozzolan, F.; Pelletier, J.; Jacquin-Joly, E.; Guerrero, A.; Maibeche-Coisne, M. Antennal esterase cDNAs from two pest moths, Spodoptera littoralis and Sesamia nonagrioides, potentially involved in odourant degradation. Insect Mol. Biol. 2007, 16, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Walker, W.B., 3rd; Roy, A.; Anderson, P.; Schlyter, F.; Hansson, B.S.; Larsson, M.C. Transcriptome Analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genom. 2019, 20, 428. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, C.A.; Teese, M.G.; Yuan, G.; Li, Y.; Scott, C.; Zhang, X.; Wu, Y.; Russell, R.J.; Oakeshott, J.G. Esterase-based metabolic resistance to insecticides in heliothine and spodopteran pests. J. Pestic. Sci. 2010, 35, 275–289. [Google Scholar] [CrossRef] [Green Version]
- El-Guindy, M.A.; Saleh, W.S.; El-Refai, A.R.A.; Abou-Donia, S.A. The role of heamolymph esterases as protectants against intoxication by fenitrothion in the cotton leafworm Spodoptera littoralis (Boisd.). Bull. Entomol. Soc. Egypt. Econ. Ser. 1984, 14, 177–197. [Google Scholar]
- Cho, J.R.; Kim, Y.J.; Kim, J.J.; Kim, H.S.; Yoo, J.K.; Lee, J.O. Electrophoretic Pattern of Larval Esterases in field and laboratory-selected strains of the tobacco cutworm, Spodoptera litura (Fabricius). J. Asia-Pac. Entomol. 1999, 2, 39–44. [Google Scholar] [CrossRef]
- Kim, Y.G.; Lee, J.I.; Kang, S.Y.; Han, S.C. Variation in insecticide susceptibilities of the beet armyworm, Spodoptera exigua (Hubner): Esterase and acetylcholinesterase activities. Korean J. Appl. Entomol. 1997, 36, 172–178. [Google Scholar]
- Yu, S.J.; Nguyen, S.N.; Abo-Elghar, G.E. Biochemical characteristics of insecticide resistance in the fall armyworm, Spodoptera frugiperda (J.E. Smith). Pestic. Biochem. Physiol. 2003, 77, 1–11. [Google Scholar] [CrossRef]
- Han, Y.; Wu, S.; Li, Y.; Liu, J.; Campbell, P.M.; Farnsworth, C.A.; Scott, C.; Russell, R.J.; Oakeshott, J.G.; Wu, Y. Proteomic and molecular analyses of esterases associated with monocrotophos resistance in Helicoverpa armigera. Pestic. Biochem. Physiol. 2012, 104, 243–251. [Google Scholar] [CrossRef]
- Li, Y.; Farnsworth, C.A.; Coppin, C.W.; Teese, M.G.; Liu, J.W.; Scott, C.; Zhang, X.; Russell, R.J.; Oakeshott, J.G. Organophosphate and pyrethroid hydrolase activities of mutant Esterases from the cotton bollworm Helicoverpa armigera. PLoS ONE 2013, 8, e77685. [Google Scholar] [CrossRef] [Green Version]
- Teese, M.G.; Farnsworth, C.A.; Li, Y.; Coppin, C.W.; Devonshire, A.L.; Scott, C.; East, P.; Russell, R.J.; Oakeshott, J.G. Heterologous expression and biochemical characterisation of fourteen esterases from Helicoverpa armigera. PLoS ONE 2013, 8, e65951. [Google Scholar] [CrossRef]
- Campbell, P.M.; Trott, J.F.; Claudianos, C.; Smyth, K.A.; Russell, R.J.; Oakeshott, J.G. Biochemistry of esterases associated with organophosphate resistance in Lucilia cuprina with comparisons to putative orthologues in other Diptera. Biochem. Genet. 1997, 35, 17–40. [Google Scholar] [CrossRef]
- Claudianos, C.; Russell, R.J.; Oakeshott, J.G. The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly. Insect Biochem. Mol. Biol. 1999, 29, 675–686. [Google Scholar] [CrossRef]
- Newcomb, R.D.; Campbell, P.M.; Ollis, D.L.; Cheah, E.; Russell, R.J.; Oakeshott, J.G. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. USA 1997, 94, 7464–7468. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Lin, Z.; Wang, H.; Liu, S.; Chang, H.; Reeck, G.; Qiao, C.; Raymond, M.; Kang, L. Two single mutations commonly cause qualitative change of nonspecific carboxylesterases in insects. Insect Biochem. Mol. Biol. 2011, 41, 1–8. [Google Scholar] [CrossRef]
- Zhu, Y.; Dowdy, A.K.; Baker, J.E. Detection of single-base substitution in an esterase gene and its linkage to malathion resistance in the parasitoid Anisopteromalus calandrae (Hymenoptera: Pteromalidae). Pestic. Sci. 1999, 55, 398–404. [Google Scholar] [CrossRef]
- Fournier, D. Mutations of acetylcholinesterase which confer insecticide resistance in insect populations. Chem. Biol. Interact. 2005, 157–158, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.C.; Callaghan, A.; Field, L.M.; Williamson, M.S.; Moores, G.D. Identification of mutations conferring insecticide-insensitive AChE in the cotton-melon aphid, Aphis gossypii Glover. Insect Mol. Biol. 2004, 13, 555–561. [Google Scholar] [CrossRef]
- Hsu, J.C.; Haymer, D.S.; Wu, W.J.; Feng, H.T. Mutations in the acetylcholinesterase gene of Bactrocera dorsalis associated with resistance to organophosphorus insecticides. Insect Biochem. Mol. Biol. 2006, 36, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Qu, M.; Denholm, I.; Fang, J.; Jiang, W.; Han, Z. Mutation in acetylcholinesterase1 associated with triazophos resistance in rice stem borer, Chilo suppressalis (Lepidoptera: Pyralidae). Biochem. Biophys. Res. Commun. 2009, 378, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Choi, J.Y.; Kim, W.T.; Je, Y.H.; Song, J.T.; Chung, B.K.; Boo, K.S.; Koh, Y.H. Mutations of acetylcholinesterase1 contribute to prothiofos-resistance in Plutella xylostella (L.). Biochem. Biophys. Res. Commun. 2007, 353, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cho, J.R.; Lee, J.I.; Kang, S.Y.; Han, S.C.; Hong, K.J.; Kim, H.S.; Yoo, J.K.; Lee, J.O. Insecticide resistance in the tobacco cutworm, Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 1998, 1, 115–122. [Google Scholar]
- Kranthi, K.R.; Jadhav, D.R.; Kranthi, S.; Wanjari, R.R.; Ali, S.S.; Russell, D.A. Insecticide resistance in five major insect pests of cotton in India. Crop. Prot. 2002, 21, 449–460. [Google Scholar] [CrossRef]
- Muthusamy, Shivakumar, Karthi, and Ramkumar, Pesticide detoxifying mechanism in field population of Spodoptera litura (Lepidoptera: Noctuidae) from South India Egypt. Acad. J. Biolog. Sci. 2011, 3, 51–57.
- Byrne, F.J.; Toscano, N.C. An insensitive acetylcholinesterase confers resistance to methomyl in the beet armyworm Spodoptera exigua (Lepidoptera: Noctuidae). J. Econ. Entomol. 2001, 94, 524–528. [Google Scholar] [CrossRef]
- Carvalho, R.A.; Omoto, C.; Field, L.M.; Williamson, M.S.; Bass, C. Investigating the molecular mechanisms of organophosphate and pyrethroid resistance in the fall armyworm Spodoptera frugiperda. PLoS ONE 2013, 8, e62268. [Google Scholar] [CrossRef] [Green Version]
- Boaventura, D.; Martin, M.; Pozzebon, A.; Mota-Sanchez, D.; Nauen, R. Monitoring of target-site mutations conferring insecticide resistance in Spodoptera frugiperda. Insects 2020, 11, 545. [Google Scholar] [CrossRef]
- Guan, F.; Zhang, J.; Shen, H.; Wang, X.; Padovan, A.; Walsh, T.K.; Tay, W.T.; Gordon, K.H.J.; James, W.; Czepak, C.; et al. Whole-genome sequencing to detect mutations associated with resistance to insecticides and Bt proteins in Spodoptera frugiperda. Insect Sci. 2020. [Google Scholar] [CrossRef]
- Jakoby, W.B.; Ziegler, D.M. The enzymes of detoxication. J. Biol. Chem. 1990, 265, 20715–20718. [Google Scholar] [CrossRef]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360 Pt 1, 1–16. [Google Scholar] [CrossRef]
- Hu, B.; Hu, S.; Huang, H.; Wei, Q.; Ren, M.; Huang, S.; Tian, X.; Su, J. Insecticides induce the co-expression of glutathione S-transferases through ROS/CncC pathway in Spodoptera exigua. Pestic. Biochem. Physiol. 2019, 155, 58–71. [Google Scholar] [CrossRef]
- Friedman, R. Genomic organization of the glutathione S-transferase family in insects. Mol. Phylogenet. Evol. 2011, 61, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Kostaropoulos, I.; Papadopoulos, A.I.; Metaxakis, A.; Boukouvala, E.; Papadopoulou-Mourkidou, E. Glutathione S-transferase in the defence against pyrethroids in insects. Insect Biochem. Mol. Biol. 2001, 31, 313–319. [Google Scholar] [CrossRef]
- Alias, Z.; Clark, A.G. Studies on the glutathione S-transferase proteome of adult Drosophila melanogaster: Responsiveness to chemical challenge. Proteomics 2007, 7, 3618–3628. [Google Scholar] [CrossRef] [PubMed]
- Lumjuan, N.; Rajatileka, S.; Changsom, D.; Wicheer, J.; Leelapat, P.; Prapanthadara, L.A.; Somboon, P.; Lycett, G.; Ranson, H. The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem. Mol. Biol. 2011, 41, 203–209. [Google Scholar] [CrossRef]
- Hardy, N.B.; Peterson, D.A.; Ross, L.; Rosenheim, J.A. Does a plant-eating insect’s diet govern the evolution of insecticide resistance? Comparative tests of the pre-adaptation hypothesis. Evol. Appl. 2018, 11, 739–747. [Google Scholar] [CrossRef] [Green Version]
- Pavlidi, N.; Vontas, J.; van Leeuwen, T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 2018, 27, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.G.; Small, G.J.; Hemingway, J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 2001, 357 Pt 1, 65–72. [Google Scholar] [CrossRef]
- Deng, H.; Huang, Y.; Feng, Q.; Zheng, S. Two epsilon glutathione S-transferase cDNAs from the common cutworm, Spodoptera litura: Characterization and developmental and induced expression by insecticides. J. Insect Physiol. 2009, 55, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Zhan, S.; Xia, X.; Xu, P.; You, H.; Jin, B.R.; Li, J. Identification and functional characterization of an epsilon glutathione S-transferase from the beet armyworm (Spodoptera exigua). Pestic. Biochem. Physiol. 2016, 132, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huang, H.; Wei, Q.; Ren, M.; Mburu, D.K.; Tian, X.; Su, J. Transcription factors CncC/Maf and AhR/ARNT coordinately regulate the expression of multiple GSTs conferring resistance to chlorpyrifos and cypermethrin in Spodoptera exigua. Pest. Manag. Sci. 2019, 75, 2009–2019. [Google Scholar] [CrossRef]
- Xu, Z.B.; Zou, X.P.; Zhang, N.; Feng, Q.L.; Zheng, S.C. Detoxification of insecticides, allechemicals and heavy metals by glutathione S-transferase SlGSTE1 in the gut of Spodoptera litura. Insect Sci. 2015, 22, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Xu, Z.; Zou, H.; Liu, J.; Chen, S.; Feng, Q.; Zheng, S. Glutathione S-transferase SlGSTE1 in Spodoptera litura may be associated with feeding adaptation of host plants. Insect Biochem Mol. Biol. 2016, 70, 32–43. [Google Scholar] [CrossRef]
- Hirowatari, A.; Chen, Z.; Mita, K.; Yamamoto, K. Enzymatic characterization of two epsilon-class glutathione S-transferases of Spodoptera litura. Arch. Insect Biochem. Physiol. 2018, 97. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Z.; Lin, X.; Feng, Q.; Zheng, S. Structure and expression of glutathione S-transferase genes from the midgut of the common cutworm, Spodoptera litura (Noctuidae) and their response to xenobiotic compounds and bacteria. J. Insect Physiol. 2011, 57, 1033–1044. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, J.; Chen, S.N.; Huang, L.H.; Feng, Q.L.; Zheng, S.C. Expression profiles of glutathione S-transferase superfamily in Spodoptera litura tolerated to sublethal doses of chlorpyrifos. Insect Sci. 2016, 23, 675–687. [Google Scholar] [CrossRef]
- Lalouette, L.; Pottier, M.A.; Wycke, M.A.; Boitard, C.; Bozzolan, F.; Maria, A.; Demondion, E.; Chertemps, T.; Lucas, P.; Renault, D.; et al. Unexpected effects of sublethal doses of insecticide on the peripheral olfactory response and sexual behavioral in a pest insect. Environ. Sci. Pollut. Res. 2016, 23, 3073–3085. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Y.L. Identification and characterisation of multiple glutathione S-transferase genes from the diamondback moth, Plutella xylostella. Pest. Manag. Sci. 2015, 71, 592–600. [Google Scholar] [CrossRef]
- Huang, H.S.; Hu, N.T.; Yao, Y.E.; Wu, C.Y.; Chiang, S.W.; Sun, C.N. Molecular cloning and heterologous expression of a glutathione S-transferase involved in insecticide resistance from the diamondback moth, Plutella xylostella. Insect Biochem. Mol. Biol. 1998, 28, 651–658. [Google Scholar] [CrossRef]
- Shi, L.; Shi, Y.; Zhang, Y.; Liao, X. A systemic study of indoxacarb resistance in Spodoptera litura revealed complex expression profiles and regulatory mechanism. Sci. Rep. 2019, 9, 14997. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Shi, Y.; Liu, M.F.; Zhang, Y.; Liao, X.L. Transcription factor CncC potentially regulates the expression of multiple detoxification genes that mediate indoxacarb resistance in Spodoptera litura. Insect Sci. 2020. [Google Scholar] [CrossRef]
- Li, Z.; Cai, T.; Qin, Y.; Zhang, Y.; Jin, R.; Mao, K.; Liao, X.; Wan, H.; Li, J. Transcriptional Response of ATP-Binding Cassette (ABC) Transporters to Insecticide in the Brown Planthopper, Nilaparvata lugens (Stal). Insects 2020, 11, 280. [Google Scholar] [CrossRef]
- Qi, W.; Ma, X.; He, W.; Chen, W.; Zou, M.; Gurr, G.M.; Vasseur, L.; You, M. Characterization and expression profiling of ATP-binding cassette transporter genes in the diamondback moth, Plutella xylostella (L.). BMC Genom. 2016, 17, 760. [Google Scholar] [CrossRef] [Green Version]
- Briz, O.; Perez-Silva, L.; Al-Abdulla, R.; Abete, L.; Reviejo, M.; Romero, M.R.; Marin, J.J.G. What “The Cancer Genome Atlas” database tells us about the role of ATP-binding cassette (ABC) proteins in chemoresistance to anticancer drugs. Expert Opin Drug Metab. Toxicol. 2019, 15, 577–593. [Google Scholar] [CrossRef] [PubMed]
- Gellatly, K.J.; Yoon, K.S.; Doherty, J.J.; Sun, W.; Pittendrigh, B.R.; Clark, J.M. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic. Biochem. Physiol. 2015, 121, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferre, J. Mechanisms of Resistance to Insecticidal Proteins from Bacillus thuringiensis. Annu. Rev. Entomol. 2021, 66, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Chen, Z.; Yang, Y.; Xiao, Y.; Liu, C.; Ma, Y.; Soberon, M.; Bravo, A.; Yang, Y.; Liu, K. A single amino acid polymorphism in ABCC2 loop 1 is responsible for differential toxicity of Bacillus thuringiensis Cry1Ac toxin in different Spodoptera (Noctuidae) species. Insect Biochem. Mol. Biol. 2018, 100, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Tanaka, S.; Adegawa, S.; Ichino, F.; Tabunoki, H.; Kikuta, S.; Sato, R. Extracellular loop structures in silkworm ABCC transporters determine their specificities for Bacillus thuringiensis Cry toxins. J. Biol. Chem. 2018, 293, 8569–8577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Maagd, R.A.; Bravo, A.; Crickmore, N. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 2001, 17, 193–199. [Google Scholar] [CrossRef]
- Banerjee, R.; Hasler, J.; Meagher, R.; Nagoshi, R.; Hietala, L.; Huang, F.; Narva, K.; Jurat-Fuentes, J.L. Mechanism and DNA-based detection of field-evolved resistance to transgenic Bt corn in fall armyworm (Spodoptera frugiperda). Sci. Rep. 2017, 7, 10877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flagel, L.; Lee, Y.W.; Wanjugi, H.; Swarup, S.; Brown, A.; Wang, J.; Kraft, E.; Greenplate, J.; Simmons, J.; Adams, N.; et al. Mutational disruption of the ABCC2 gene in fall armyworm, Spodoptera frugiperda, confers resistance to the Cry1Fa and Cry1A.105 insecticidal proteins. Sci. Rep. 2018, 8, 7255. [Google Scholar] [CrossRef] [Green Version]
- Boaventura, D.; Ulrich, J.; Lueke, B.; Bolzan, A.; Okuma, D.; Gutbrod, O.; Geibel, S.; Zeng, Q.; Dourado, P.M.; Martinelli, S.; et al. Molecular characterization of Cry1F resistance in fall armyworm, Spodoptera frugiperda from Brazil. Insect Biochem. Mol. Biol. 2020, 116, 103280. [Google Scholar] [CrossRef]
- Park, Y.; Gonzalez-Martinez, R.M.; Navarro-Cerrillo, G.; Chakroun, M.; Kim, Y.; Ziarsolo, P.; Blanca, J.; Canizares, J.; Ferre, J.; Herrero, S. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biol. 2014, 12, 46. [Google Scholar] [CrossRef] [Green Version]
- Moar, W.J.; Pusztai-Carey, M.; van Faassen, H.; Bosch, D.; Frutos, R.; Rang, C.; Luo, K.; Adang, M.J. Development of Bacillus thuringiensis CryIC resistance by Spodoptera exigua (Hubner) (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 1995, 61, 2086–2092. [Google Scholar] [CrossRef] [Green Version]
- Müller-Cohn, J.; Chaufaux, J.; Buisson, C.; Gilois, N.; Sanchis, V.; Lereclus, D. Spodoptera littoralis (Lepidoptera: Noctuidae) resistance to CryIC and cross-resistance to other Bacillus thuringiensis crystal toxins. J. Econ. Entomol. 1996, 89, 791–797. [Google Scholar] [CrossRef]
- Barkhade, U.P.; Thakare, A.S. Protease mediated resistance mechanism to Cry1C and Vip3A in Spodoptera litura. Egypt. Acad. J. Biol. Sci. 2010, 6, 43–50. [Google Scholar] [CrossRef]
- Hernandez-Martinez, P.; Navarro-Cerrillo, G.; Caccia, S.; de Maagd, R.A.; Moar, W.J.; Ferre, J.; Escriche, B.; Herrero, S. Constitutive activation of the midgut response to Bacillus thuringiensis in Bt-resistant Spodoptera exigua. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Pinos, D.; Martinez-Solis, M.; Herrero, S.; Ferre, J.; Hernandez-Martinez, P. The Spodoptera exigua ABCC2 acts as a Cry1A receptor independently of its nucleotide binding domain II. Toxins 2019, 11, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Xu, Y.; Zuo, Y.; Yang, Y.; Tabashnik, B.E.; Wu, Y. Evaluation of five candidate receptors for three Bt toxins in the beet armyworm using CRISPR-mediated gene knockouts. Insect Biochem. Mol. Biol. 2020, 121, 103361. [Google Scholar] [CrossRef]
- Abdelgaffar, H.; Perera, O.P.; Jurat-Fuentes, J.L. ABC transporter mutations in Cry1F-resistant fall armyworm (Spodoptera frugiperda) do not result in altered susceptibility to selected small molecule pesticides. Pest. Manag. Sci. 2021, 77, 949–955. [Google Scholar] [CrossRef]
- Xiang, M.; Zhang, L.; Lu, Y.; Tang, Q.; Liang, P.; Shi, X.; Song, D.; Gao, X. A P-glycoprotein gene serves as a component of the protective mechanisms against 2-tridecanone and abamectin in Helicoverpa armigera. Gene 2017, 627, 63–71. [Google Scholar] [CrossRef]
- Jin, M.; Liao, C.; Chakrabarty, S.; Zheng, W.; Wu, K.; Xiao, Y. Transcriptional response of ATP-binding cassette (ABC) transporters to insecticides in the cotton bollworm, Helicoverpa armigera. Pestic. Biochem. Physiol. 2019, 154, 46–59. [Google Scholar] [CrossRef]
- Zuo, Y.Y.; Huang, J.L.; Wang, J.; Feng, Y.; Han, T.T.; Wu, Y.D.; Yang, Y.H. Knockout of a P-glycoprotein gene increases susceptibility to abamectin and emamectin benzoate in Spodoptera exigua. Insect Mol. Biol. 2017. [Google Scholar] [CrossRef]
- Jin, M.; Yang, Y.; Shan, Y.; Chakrabarty, S.; Cheng, Y.; Soberon, M.; Bravo, A.; Liu, K.; Wu, K.; Xiao, Y. Two ABC transporters are differentially involved in the toxicity of two Bacillus thuringiensis Cry1 toxins to the invasive crop-pest Spodoptera frugiperda (J. E. Smith). Pest. Manag. Sci. 2021, 77, 1492–1501. [Google Scholar] [CrossRef]
- Genovese, I.; Ilari, A.; Assaraf, Y.G.; Fazi, F.; Colotti, G. Not only P-glycoprotein: Amplification of the ABCB1-containing chromosome region 7q21 confers multidrug resistance upon cancer cells by coordinated overexpression of an assortment of resistance-related proteins. Drug Resist. Updat. 2017, 32, 23–46. [Google Scholar] [CrossRef]
- Zhu, B.; Sun, X.; Nie, X.; Liang, P.; Gao, X. MicroRNA-998-3p contributes to Cry1Ac-resistance by targeting ABCC2 in lepidopteran insects. Insect Biochem. Mol. Biol. 2020, 117, 103283. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, Y.; Yuan, W.; Xiao, Y.; Liu, C.; Wang, J.; Peng, J.; Peng, R.; Soberon, M.; Bravo, A.; et al. FOXA transcriptional factor modulates insect susceptibility to Bacillus thuringiensis Cry1Ac toxin by regulating the expression of toxin-receptor ABCC2 and ABCC3 genes. Insect Biochem. Mol. Biol. 2017, 88, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bo, H.; Miaomiao, R.; Jianfeng, F.; Sufang, H.; Xia, W.; Elzaki, M.E.A.; Chris, B.; Palli, S.R.; Jianya, S. Xenobiotic transcription factors CncC and maf regulate expression of CYP321A16 and CYP332A1 that mediate chlorpyrifos resistance in Spodoptera exigua. J. Hazard. Mater. 2020, 398, 122971. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zeng, X.; Wen, S.; Liu, X.; Shang, Q. Characterization of the Cap ‘n’ Collar Isoform C gene in Spodoptera frugiperda and its Association with Superoxide Dismutase. Insects 2020, 11, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Insecticide Chemical Class | S. exigua | S. frugiperda | S. littoralis | S. litura |
|---|---|---|---|---|
| Avermectins | Abamectin (#31), emamectin benzoate (#48) | Abamectin (#43), emamectin benzoate (#27) | ||
| Benzoylurea | Chlorfluazuron (#9), diflubenzuron, lufenuron (#10), teflubenzuron | Lufenuron (#2), triflumuron | Diflubenzuron, teflubenzuron (#3) | Lufenuron (#8) |
| Bacillus thuringensis | Bt var. unspecified (#10), var aizawai, var. kurstaki | Bt var unspecified (#3), var aizawai | ||
| Bt toxins | Cry1Ca | Cry1Aa, Cry1A.105, Cry1Ab (#2), Cry1Ac (#3), Cry1F (#54), Cry2Ab2 (#2), Vip3A (#3) | ||
| Carbamates | Methomyl (#18), thiodicarb | Carbaryl (#7), methomyl (#6), thiodicarb (#2) | Carbaryl (#2), methomyl (#2), | Carbaryl (#2), methomyl (#38), thiodicarb (#33) |
| Cyclodienes | BHC/cyclodiene—unspecified in literature (#3), endosulfan (#19) | Aldrin, dieldrin, lindane (#2) | Endrin, toxaphene (#2) | Endosulfan (#31), lindane |
| Diamides | Chlorantraniliprole (#26), cyantraniliprole, flubendiamide (#5) | Chlorantraniliprole (#2), flubendiamide (#2) | Chlorantraniliprole (#11) | |
| Diacylhydrazines | Methoxyfenozide (#25), tebufenozide (#19) | Tebufenozide | Methoxyfenozide (#36), tebufenozide | |
| Neonicotinoids | Acetamiprid | |||
| Organochlorine | DDT (#4) | DDT (#3) | DDT | DDT (#2) |
| Organophosphates | Chlorpyrifos (#48), parathion-methyl (#3), profenofos (#22), quinalphos (#9) | Acephate, chlorpurifos (#7), diazinon (#2), dichlorvos, malathion (#2), parathion-methyl (#4), sulprofos, trichlorfon, | Acephate, azinphos-methyl, chlorpyrifos (#4), fenitrothion, leptophos, methamidophos, methidathion, monocrotophos (#4), parathion (#2), parathion-methyl (#3), profenofos, sulprofos, triazophos, trichlorfon | Chlorfenvinphos, chlorpyrifos (#55), diazinon, dichlorvos, malathion, monocrotophos (#5), phoxim (#4), profenofos (#56), quinalphos (#9), triazophos (#2), trichlorfon |
| Oxadiazines | Indoxacarb (#44), metaflumizone (#4) | Indoxacarb | Indoxacarb (#50), metaflumizone | |
| Phenylpyrazoles | Fipronil (#15) | |||
| Pyrethroids | Bifenthrine (#13), cyfluthrin (#2), cyhalothrin-lambda (#2), cypermethrin (#56), cypermethrin-beta, deltamethrin (#43), fenpropathrin (#14), fenvalerate (#5), permethrin (#2), pyrethroids-unspecified in literature (#3) | Bifenthrine, cyfluthrin, cyhalothrin, cyhalothrin-lambda (#11), cypermethrin (#2), cypermethrin-zeta, deltamethrin (#2), fenvalerate, fluvalinate, permethrin (#5), tau-fluvalinate, tralomethrin | Cypermethrin (#4), deltamethrin (#2), fenvalerate, flucythrinate, permethrin | Bifenthrine (#33), cyfluthrin (#12), cyfluthrin-beta (#9), cyhalothrin (#3), cyhalothrin-lambda (#11), cypermethrin (#49), cypermethrin-beta (#15), deltamethrin (#50), esfenvalerate (#9), fenpropathrin (#10), fenvalerate (#7), pyrethrins |
| Pyrroles | Chlorfenapyr (#11) | Chlorfenapyr (#5) | ||
| Spinosyns | Spinetoram, spinosad (#56) | Spinetoram (#2), spinosad (#2) | Spinosad (#39) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hilliou, F.; Chertemps, T.; Maïbèche, M.; Le Goff, G. Resistance in the Genus Spodoptera: Key Insect Detoxification Genes. Insects 2021, 12, 544. https://doi.org/10.3390/insects12060544
Hilliou F, Chertemps T, Maïbèche M, Le Goff G. Resistance in the Genus Spodoptera: Key Insect Detoxification Genes. Insects. 2021; 12(6):544. https://doi.org/10.3390/insects12060544
Chicago/Turabian StyleHilliou, Frédérique, Thomas Chertemps, Martine Maïbèche, and Gaëlle Le Goff. 2021. "Resistance in the Genus Spodoptera: Key Insect Detoxification Genes" Insects 12, no. 6: 544. https://doi.org/10.3390/insects12060544