Indirect Volume Estimation for Acute Ischemic Stroke from Diffusion Weighted Image Using Slice Image Segmentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Imaging Acquisition
2.3. Annotation
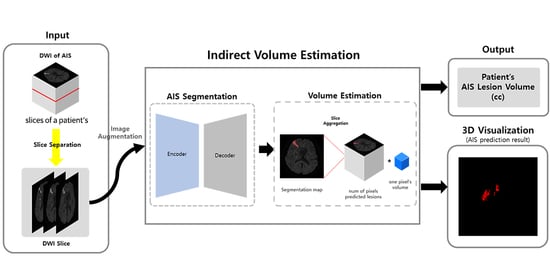
2.4. Method
2.4.1. Augmentation
2.4.2. Volume Estimation
- Direct Volume Estimation (Using 3D Segmentation)
- Indirect Volume Estimation (Using 2D Segmentation)
2.4.3. Loss Function
2.4.4. Evaluation Metrics
3. Results
3.1. Implementation Details
3.2. Segmentation Performance
3.3. Volume Estimation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Emberson, J.; Lees, K.R.; Lyden, P.; Blackwell, L.; Albers, G.; Bluhmki, E.; Brott, T.; Cohen, G.; Davis, S.; Donnan, G.; et al. Effect of Treatment Delay, Age, and Stroke Severity on the Effects of Intravenous Thrombolysis with Alteplase for Acute Ischaemic Stroke: A Meta-Analysis of Individual Patient Data from Randomised Trials. Lancet 2014, 384, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Lopez, A.D.; Mathers, C.D.; Ezzati, M.; Jamison, D.T.; Murray, C.J.L. Global and Regional Burden of Disease and Risk Factors, 2001: Systematic Analysis of Population Health Data. Lancet 2006, 367, 1747–1757. [Google Scholar] [CrossRef]
- Saver, J.L.; Goyal, M.; van der Lugt, A.; Menon, B.K.; Majoie, C.B.L.M.; Dippel, D.W.; Campbell, B.C.; Nogueira, R.G.; Demchuk, A.M.; Tomasello, A.; et al. Time to Treatment with Endovascular Thrombectomy and Outcomes from Ischemic Stroke: A Meta-Analysis. JAMA 2016, 316, 1279–1289. [Google Scholar] [CrossRef]
- Ma, H.; Campbell, B.C.V.; Parsons, M.W.; Churilov, L.; Levi, C.R.; Hsu, C.; Kleinig, T.J.; Wijeratne, T.; Curtze, S.; Dewey, H.M.; et al. Thrombolysis Guided by Perfusion Imaging up to 9 Hours after Onset of Stroke. N. Engl. J. Med. 2019, 380, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Albers, G.W.; Marks, M.P.; Kemp, S.; Christensen, S.; Tsai, J.P.; Ortega-Gutierrez, S.; McTaggart, R.A.; Torbey, M.T.; Kim-Tenser, M.; Leslie-Mazwi, T.; et al. Thrombectomy for Stroke at 6 to 16 Hours with Selection by Perfusion Imaging. N. Engl. J. Med. 2018, 378, 708–718. [Google Scholar] [CrossRef]
- Nogueira, R.G.; Jadhav, A.P.; Haussen, D.C.; Bonafe, A.; Budzik, R.F.; Bhuva, P.; Yavagal, D.R.; Ribo, M.; Cognard, C.; Hanel, R.A.; et al. Thrombectomy 6 to 24 Hours after Stroke with a Mismatch between Deficit and Infarct. N. Engl. J. Med. 2018, 378, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Edlow, B.L.; Hurwitz, S.; Edlow, J.A. Diagnosis of DWI-Negative Acute Ischemic Stroke: A Meta-Analysis. Neurology 2017, 89, 256–262. [Google Scholar] [CrossRef]
- Muir, K.W.; Buchan, A.; von Kummer, R.; Rother, J.; Baron, J.-C. Imaging of Acute Stroke. Lancet Neurol. 2006, 5, 755–768. [Google Scholar] [CrossRef]
- Wheeler, H.M.; Mlynash, M.; Inoue, M.; Tipirneni, A.; Liggins, J.; Zaharchuk, G.; Straka, M.; Kemp, S.; Bammer, R.; Lansberg, M.G.; et al. Early Diffusion-Weighted Imaging and Perfusion-Weighted Imaging Lesion Volumes Forecast Final Infarct Size in DEFUSE 2. Stroke 2013, 44, 681–685. [Google Scholar] [CrossRef]
- Mitra, J.; Bourgeat, P.; Fripp, J.; Ghose, S.; Rose, S.; Salvado, O.; Connelly, A.; Campbell, B.; Palmer, S.; Sharma, G.; et al. Lesion Segmentation from Multimodal MRI Using Random Forest Following Ischemic Stroke. NeuroImage 2014, 98, 324–335. [Google Scholar] [CrossRef]
- Maier, O.; Schröder, C.; Forkert, N.D.; Martinetz, T.; Handels, H. Classifiers for Ischemic Stroke Lesion Segmentation: A Comparison Study. PLoS ONE 2015, 10, e0145118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Zhao, L.; Lou, W.; Abrigo, J.M.; Mok, V.C.T.; Chu, W.C.W.; Wang, D.; Shi, L. Automatic Segmentation of Acute Ischemic Stroke From DWI Using 3-D Fully Convolutional DenseNets. IEEE Trans. Med. Imaging 2018, 37, 2149–2160. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, F.; Sparks, R.; Ourselin, S. TorchIO: A Python Library for Efficient Loading, Preprocessing, Augmentation and Patch-Based Sampling of Medical Images in Deep Learning. Comput. Methods Programs Biomed. 2021, 208, 106236. [Google Scholar] [CrossRef] [PubMed]
- Buslaev, A.; Parinov, A.; Khvedchenya, E.; Iglovikov, V.I.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef] [Green Version]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. arXiv 2016, arXiv:160606650. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:150504597. [Google Scholar]
- Buda, M.; Saha, A.; Mazurowski, M.A. Association of Genomic Subtypes of Lower-Grade Gliomas with Shape Features Automatically Extracted by a Deep Learning Algorithm. Comput. Biol. Med. 2019, 109, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Taha, A.A.; Hanbury, A. Metrics for Evaluating 3D Medical Image Segmentation: Analysis, Selection, and Tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [Green Version]
- Ellis, D.G.; Aizenberg, M.R. Deep Learning Using Augmentation via Registration: 1st Place Solution to the AutoImplant 2020 Challenge. In Towards the Automatization of Cranial Implant Design in Cranioplasty, Proceedings of the 23rd International Conference, Lima, Peru, 4–8 October 2020; Li, J., Egger, J., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 47–55. [Google Scholar]
- Zettler, N.; Mastmeyer, A. Comparison of 2D vs. 3D U-Net Organ Segmentation in Abdominal 3D CT Images. arXiv 2021, arXiv:210704062. [Google Scholar]
- Barber, P.A.; Hill, M.D.; Eliasziw, M.; Demchuk, A.M.; Pexman, J.H.W.; Hudon, M.E.; Tomanek, A.; Frayne, R.; Buchan, A.M. ASPECTS Study Group Imaging of the Brain in Acute Ischaemic Stroke: Comparison of Computed Tomography and Magnetic Resonance Diffusion-Weighted Imaging. J. Neurol. Neurosurg. Psychiatry 2005, 76, 1528–1533. [Google Scholar] [CrossRef]
- Powers, W.J.; Rabinstein, A.A.; Ackerson, T.; Adeoye, O.M.; Bambakidis, N.C.; Becker, K.; Biller, J.; Brown, M.; Demaerschalk, B.M.; Hoh, B.; et al. 2018 Guidelines for the Early Management of Patients with Acute Ischemic Stroke: A Guideline for Healthcare Professionals from the American Heart Association/American Stroke Association. Stroke 2018, 49, e46–e99. [Google Scholar] [CrossRef] [PubMed]
- Saver, J.L. Time Is Brain—Quantified. Stroke 2006, 37, 263–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahlgren, N.; Moreira, T.; Michel, P.; Steiner, T.; Jansen, O.; Cognard, C.; Mattle, H.P.; van Zwam, W.; Holmin, S.; Tatlisumak, T.; et al. Mechanical Thrombectomy in Acute Ischemic Stroke: Consensus Statement by ESO-Karolinska Stroke Update 2014/2015, Supported by ESO, ESMINT, ESNR and EAN. Int. J. Stroke Off. J. Int. Stroke Soc. 2016, 11, 134–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Model | Sensitivity (%) | Specificity (%) | F1-Score (%) | Jaccard Index (%) | |
|---|---|---|---|---|---|
| Direct | Scratch | 75.27 | 48.02 | 54.76 | 42.02 |
| Pretrained (Auto Implant) | 71.00 | 49.31 | 55.71 | 43.04 | |
| Pretrained (Tumor) | 63.72 | 52.27 | 52.48 | 39.88 | |
| Indirect | Scratch | 71.11 | 75.87 | 73.09 | 58.49 |
| Pretrained (Glioma) | 75.0 | 77.87 | 76.02 | 62.12 | |
| Model | Sensitivity (%) | Specificity (%) | F1-Score (%) | Jaccard Index (%) | |
|---|---|---|---|---|---|
| Direct | Scratch | 70.99 | 63.66 | 63.94 | 51.48 |
| Pretrained (Auto Implant) | 63.22 | 58.65 | 56.73 | 44.93 | |
| Pretrained (Tumor) | 67.05 | 62.26 | 60.33 | 48.33 | |
| Indirect | Scratch | 69.35 | 83.75 | 73.93 | 60.28 |
| Pretrained (Glioma) | 72.81 | 84.33 | 77.23 | 63.82 | |
| Model | VS (%) | MAE (cc) | |
|---|---|---|---|
| External validation | Direct | 62.59 | 5.706 |
| Indirect | 89.17 | 2.468 | |
| Internal validation | Direct | 67.68 | 1.159 |
| Indirect | 93.25 | 0.797 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-A.; Jang, J.-W.; Park, S.-W.; Kim, P.-J.; Yeo, N.-Y.; Kim, C.; Kim, Y.; Choi, H.-S.; Kim, S. Indirect Volume Estimation for Acute Ischemic Stroke from Diffusion Weighted Image Using Slice Image Segmentation. J. Pers. Med. 2022, 12, 521. https://doi.org/10.3390/jpm12040521
Lee S-A, Jang J-W, Park S-W, Kim P-J, Yeo N-Y, Kim C, Kim Y, Choi H-S, Kim S. Indirect Volume Estimation for Acute Ischemic Stroke from Diffusion Weighted Image Using Slice Image Segmentation. Journal of Personalized Medicine. 2022; 12(4):521. https://doi.org/10.3390/jpm12040521
Chicago/Turabian StyleLee, Seung-Ah, Jae-Won Jang, Sang-Won Park, Pum-Jun Kim, Na-Young Yeo, Chulho Kim, Yoon Kim, Hyun-Soo Choi, and Seongheon Kim. 2022. "Indirect Volume Estimation for Acute Ischemic Stroke from Diffusion Weighted Image Using Slice Image Segmentation" Journal of Personalized Medicine 12, no. 4: 521. https://doi.org/10.3390/jpm12040521