Cytotaxonomy and Molecular Analyses of Mycteria americana (Ciconiidae: Ciconiiformes): Insights on Stork Phylogeny
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples, Cell Culture, and Chromosome Preparations
2.2. Classical Cytogenetics Experiments
2.3. Fluorescent In Situ Hybridization (FISH) Experiments
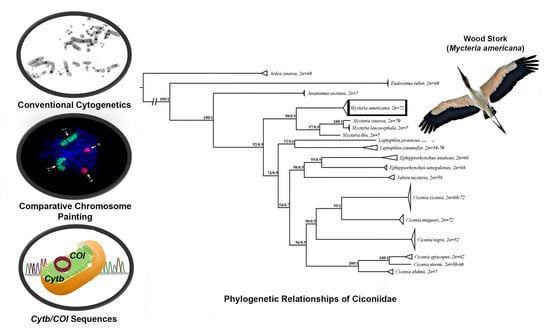
2.4. Molecular Phylogenetic Analysis
3. Results
3.1. Karyotype Characterization
3.2. Chromosome Painting
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guzzi, A.; Santos-Nascimento, M.; Lima, S.P.; Santos, S.S.; Donatelli, R.J. Osteologia craniana e aspectos evolutivos de Mycteria (Aves: Ciconiidae). Rev. Nord. Biol. 2014, 23, 85–103. [Google Scholar]
- Donatelli, R.J.; Guzzi, A.; Nobushige, S.Y.L.; Ribeiro, A.S.N.; Santos, S.S.; Ferreira, G.J.B.D.C.; Santos, F.D.C.V. Phylogeny of the species of Ciconia (Aves, Ciconiidae) based on cranial osteological characteristics. Comun. Sci. 2019, 9, 575–589. [Google Scholar]
- Hackett, S.J.; Kimball, R.T.; Reddy, S.; Bowie, R.C.K.; Braun, E.L.; Braun, M.J.; Chojnowski, J.L.; Cox, W.A.; Han, K.-L.; Harshman, J.; et al. A Phylogenomic Study of Birds Reveals Their Evolutionary History. Science 2008, 320, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Gibb, G.C.; Kennedy, M.; Penny, D. Beyond phylogeny: Pelecaniform and ciconiiform birds, and long-term niche stability. Mol. Phylogenetics Evol. 2013, 68, 229–238. [Google Scholar] [CrossRef]
- Kimball, R.T.; Wang, N.; Heimer-McGinn, V.; Ferguson, C.; Braun, E.L. Identifying localized biases in large datasets: A case study using the avian tree of life. Mol. Phylogenetics Evol. 2013, 69, 1021–1032. [Google Scholar] [CrossRef]
- Kuramoto, T.; Nishihara, H.; Watanabe, M.; Okada, N. Determining the Position of Storks on the Phylogenetic Tree of Waterbirds by Retroposon Insertion Analysis. Genome Biol. Evol. 2015, 7, 3180–3189. [Google Scholar] [CrossRef]
- Gill, F.; Donsker, D.; Rasmussen, P. IOC World Bird List (v13.1). 2023. Available online: https://www.worldbirdnames.org/new/ (accessed on 5 January 2023).
- Seligmann, I.C.; Furo, I.O.; Santos, M.S.; Tagliarini, M.M.; Araujo, C.C.; Ferguson-Smith, M.A.; Oliveira, E.H. Comparative chromosome painting in two Brazilian stork species with different diploid numbers. Cytogenet. Genome Res. 2019, 159, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Kahl, M.P. Family Ciconiidae. In Check-List of Birds of the World, 2nd ed.; Mayr, E., Cottrell, G.W., Eds.; Museum of Comparative Zoology: Cambridge, MA, USA, 1979; pp. 245–252. [Google Scholar]
- Wood, D.S. Concordance between classifications of the Ciconiidae based on behavioral and morphological data. J. Ornithol. 1984, 125, 25–37. [Google Scholar] [CrossRef]
- Slikas, B. Phylogeny of the Avian Family Ciconiidae (Storks) Based on Cytochrome b Sequences and DNA–DNA Hybridization Distances. Mol. Phylogenetics Evol. 1997, 8, 275–300. [Google Scholar] [CrossRef]
- Slikas, B. Recognizing and testing homology of courtship displays in storks (Aves: Ciconiiformes: Ciconiidae). Evolution 1998, 52, 884–893. [Google Scholar] [CrossRef]
- De Pietri, V.L.; Mayr, G. The phylogenetic relationships of the Early Miocene stork Grallavis edwardsi, with comments on the interrelationships of living Ciconiidae (Aves). Zool. Scr. 2014, 43, 576–585. [Google Scholar] [CrossRef]
- Santos, S.S.; Nobushige, S.Y.L.; Ribeiro, A.S.N.; Santos, F.D.C.V.; Donatelli, R.J.; Ferreira, G.J.B.D.C.; Guzzi, A. Phylogeny of the species of Ciconia (Aves, Ciconiidae) based on cranial osteological characteristics. Comun. Sci. 2018, 9, 575–589. [Google Scholar] [CrossRef]
- Belterman, R.H.R.; De Boer, L.E.M. A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica 1984, 65, 39–82. [Google Scholar] [CrossRef]
- Belterman, R.H.R.; De Boer, L.E.M. A miscellaneous collection of bird karyotypes. Genetica 1990, 83, 17–29. [Google Scholar] [CrossRef]
- BirdLife International. Endemic Bird Areas factsheet: Cameroon mountains. Available online: http://www.birdlife.org (accessed on 6 January 2023).
- Riojas-López, M.E.; Mellink, E. A New Wood Stork (Mycteria americana) Colony in Western Mexico. Waterbirds Int. J. Waterbird Biol. 2016, 104–107. [Google Scholar] [CrossRef]
- Lopes, I.; Miño, C.; Del Lama, S. Genetic diversity and evidence of recent demographic expansion in waterbird populations from the Brazilian Pantanal. Braz. J. Biol. 2007, 67, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Lopes, I.; Tomasulo-Seccomandi, A.; Bryan, A., Jr.; Brisbin, I., Jr.; Glenn, T.; Del Lama, S. Genetic status of the wood stork (Mycteria americana) from the southeastern United States and the Brazilian Pantanal as revealed by mitochondrial DNA analysis. Genet. Mol. Res. 2011, 10, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Llanes–Quevedo, A.; González, M.A.; Mena, R.C.; Frankel, C.; Lopez, G.E. Microsatellite variability of the wood stork Mycteria americana (Aves, Ciconidae) in Cuba: Implications for its conservation. Anim. Biodivers. Conserv. 2018, 41, 357–364. [Google Scholar] [CrossRef]
- Francisco, M.R.; Galetti-Jr, P.M. First karyotypical description of two American Ciconiiform birds, Mycteria americana (Ciconiidae) and Platalea ajaja (Threskiornithidae) and its significance for the chromosome evolutionary and biological conservation approaches. Genet. Mol. Biol. 2000, 23, 799–801. [Google Scholar] [CrossRef]
- de Oliveira Furo, I.; Kretschmer, R.; O’Brien, P.C.; Ferguson-Smith, M.A.; de Oliveira, E.H.C. Chromosomal Diversity and Karyotype Evolution in South American Macaws (Psittaciformes, Psittacidae). PLoS ONE 2015, 10, e0130157. [Google Scholar] [CrossRef]
- Kretschmer, R.; Gunski, R.J.; Garnero, A.D.V.; De Freitas, T.R.O.; Toma, G.A.; Cioffi, M.D.B.; De Oliveira, E.H.C.; O’Connor, R.E.; Griffin, D.K. Chromosomal Analysis in Crotophaga ani (Aves, Cuculiformes) Reveals Extensive Genomic Reorganization and an Unusual Z-Autosome Robertsonian Translocation. Cells 2020, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, W.; Hu, Y.; Li, S.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Nie, W. Comparative chromosome maps between the stone curlew and three ciconiiform species (the grey heron, little egret and crested ibis). BMC Ecol. Evol. 2022, 22, 1–13. [Google Scholar] [CrossRef]
- Sasaki, M.; Ikeuchi, T.; Makino, S. A feather pulp culture technique for avian chromosomes, with notes on the chromosomes of the peafowl and the ostrich. Cell. Mol. Life Sci. 1968, 24, 1292–1293. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.S. Reviewing the chromosome nomenclature of Levan et al. Brazil. J. Genet. 1987, 9, 741–743. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, E.H.C.; Tagliarini, M.M.; Rissino, J.D.; Pieczarka, J.C.; Nagamachi, C.Y.; O’Brien, P.C.M.; Ferguson-Smith, M.A. Reciprocal chromosome painting between white hawk (Leucopternis albicollis) and chicken reveals extensive fusions and fissions during karyotype evolution of accipitridae (Aves, Falconiformes). Chromosom. Res. 2010, 18, 349–355. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 27 December 2022).
- Abu Shnaf, A.S.M.; Al-Khalifa, M.S. First constitutive heterochromatin characterization and Karyotype of white stork Ciconia ciconia (Aves: Ciconiidae). Braz. J. Biol. 2023, 83. [Google Scholar] [CrossRef]
- Kretschmer, R.; de Oliveira, E.H.C.; Dos Santos, M.S.; Furo, I.D.O.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Garnero, A.D.V.; Gunski, R.J. Chromosome mapping of the large elaenia (Elaenia spectabilis): Evidence for a cytogenetic signature for passeriform birds? Biol. J. Linn. Soc. 2015, 115, 391–398. [Google Scholar] [CrossRef]
- Gunski, R.J.; Kretschmer, R.; Souza, M.S.; Oliveira-Furo, I.; Barcellos, S.A.; Costa, A.L.; Valle Garnero, A. Evolution of bird sex chromosomes narrated by repetitive sequences: Unusual W chromosome enlargement in Gallinula melanops (Aves: Grui-formes: Rallidae). Cytogenet. Genome Res. 2019, 158, 152–159. [Google Scholar] [CrossRef]
- Furo, I.D.O.; Kretschmer, R.; O’Brien, P.C.M.; Pereira, J.C.; Ferguson-Smith, M.A.; de Oliveira, E.H.C. Phylogenetic Analysis and Karyotype Evolution in Two Species of Core Gruiformes: Aramides cajaneus and Psophia viridis. Genes 2020, 11, 307. [Google Scholar] [CrossRef]
- Kretschmer, R.; Gunski, R.J.; Garnero, A.; de Oliveira Furo, I.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; de Oliveira, E.H.C. Molecular Cytogenetic Characterization of Multiple Intrachromosomal Rearrangements in Two Representatives of the Genus Turdus (Turdidae, Passeriformes). PLoS ONE 2014, 9, e103338. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, M.D.S.; Kretschmer, R.; Frankl-Vilches, C.; Bakker, A.; Gahr, M.; O´brien, P.C.M.; Ferguson-Smith, M.A.; de Oliveira, E.H.C. Comparative Cytogenetics between Two Important Songbird, Models: The Zebra Finch and the Canary. PLoS ONE 2017, 12, e0170997. [Google Scholar] [CrossRef]
- Kretschmer, R.; de Souza, M.; Furo, I.; Romanov, M.; Gunski, R.; Garnero, A.; de Freitas, T.; de Oliveira, E.; O’Connor, R.; Griffin, D. Interspecies Chromosome Mapping in Caprimulgiformes, Piciformes, Suliformes, and Trogoniformes (Aves): Cytogenomic Insight into Microchromosome Organization and Karyotype Evolution in Birds. Cells 2021, 10, 826. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sousa, R.P.C.; Campos, P.S.B.; dos Santos, M.d.S.; O’Brien, P.C.; Ferguson-Smith, M.A.; de Oliveira, E.H.C. Cytotaxonomy and Molecular Analyses of Mycteria americana (Ciconiidae: Ciconiiformes): Insights on Stork Phylogeny. Genes 2023, 14, 816. https://doi.org/10.3390/genes14040816
de Sousa RPC, Campos PSB, dos Santos MdS, O’Brien PC, Ferguson-Smith MA, de Oliveira EHC. Cytotaxonomy and Molecular Analyses of Mycteria americana (Ciconiidae: Ciconiiformes): Insights on Stork Phylogeny. Genes. 2023; 14(4):816. https://doi.org/10.3390/genes14040816
Chicago/Turabian Stylede Sousa, Rodrigo Petry Corrêa, Paula Sabrina Bronze Campos, Michelly da Silva dos Santos, Patricia Caroline O’Brien, Malcolm Andrew Ferguson-Smith, and Edivaldo Herculano Corrêa de Oliveira. 2023. "Cytotaxonomy and Molecular Analyses of Mycteria americana (Ciconiidae: Ciconiiformes): Insights on Stork Phylogeny" Genes 14, no. 4: 816. https://doi.org/10.3390/genes14040816