Current Approaches to and the Application of Intracytoplasmic Sperm Injection (ICSI) for Avian Genome Editing
Abstract
:1. Introduction
2. CRISPR/Cas9-Mediated Genome Editing Technology
3. Current Approaches for Avian Genome Editing Based on the CRISPR/Cas9 System
3.1. Viral Infection
3.2. Chimeric Method Using PGCs
3.3. Sperm-Mediated Genome Editing
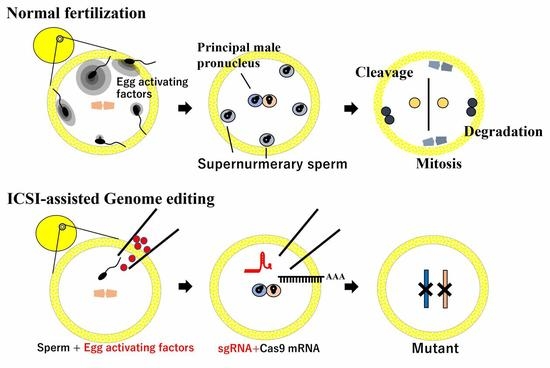
4. ICSI-Assisted Genome Editing
4.1. Establishment of ICSI
4.2. ICSI-Assisted Genome Editing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, G.; Han, J.Y. Avian biomodels for use as pharmaceutical bioreactors and for studying human diseases. Ann. N. Y. Acad. Sci. 2011, 1229, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Sipp, D.; Enomoto, H. Tissue interactions in neural crest cell development and disease. Science 2013, 341, 860–863. [Google Scholar] [CrossRef] [PubMed]
- Naito, M. Development of avian embryo manipulation techniques and their application to germ cell manipulation. Anim. Sci. J. 2003, 74, 157–168. [Google Scholar] [CrossRef]
- Han, J.Y. Germ cells and transgenesis in chickens. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Hillier, L.W.; Miller, W.; Birney, E.; Warren, W.; Hardison, R.C.; Ponting, C.P.; Bork, P.; Burt, D.W.; Groenen, M.A.M.; Delany, M.E.; et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef] [Green Version]
- Kawahara-Miki, R.; Sano, S.; Nunome, M.; Shimmura, T.; Kuwayama, T.; Takahashi, S.; Kawashima, T.; Matsuda, Y.; Yoshimura, T.; Kono, T. Next-generation sequencing reveals genomic features in the Japanese quail. Genomics 2013, 101, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Dalloul, R.A.; Long, J.A.; Zimin, A.V.; Aslam, L.; Beal, K.; Blomberg, L.A.; Bouffard, P.; Burt, S.W.; Crasta, O.; Croojimans, R.P.; et al. Multiplatform next-generation sequencing of the domestic turkey (Meleagris gallopavo): Genome assembly and analysis. PLoS Biol. 2010, 8, e1000475. [Google Scholar] [CrossRef]
- Warren, W.C.; Clayton, D.F.; Ellegren, H.; Arnold, A.P.; Hillier, L.W.; Künstner, A.; Searle, S.; White, S.; Vilella, A.J.; Fairley, S.; et al. The genome of a songbird. Nature 2010, 464, 757–762. [Google Scholar] [CrossRef] [Green Version]
- Schusser, B.; Collarini, E.J.; Yi, H.; Izquierdo, S.M.; Fesler, J.; Pedersen, D.; Klasing, K.C.; Kaspers, B.; Harriman, W.D.; van de Lavoir, M.C.; et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. USA 2013, 110, 20170–20175. [Google Scholar] [CrossRef] [Green Version]
- Thomas, K.R.; Capecchi, M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987, 51, 503–512. [Google Scholar] [CrossRef]
- Schusser, B.; Collarini, E.J.; Pedersen, D.; Yi, H.; Ching, K.; Izquierdo, S.; Thoma, T.; Lettmann, S.; Kaspers, B.; Etches, R.J.; et al. Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur. J. Immunol. 2016, 46, 2137–2148. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Kang, K.S.; Han, J.Y. Current genomic editing approaches in avian transgenesis. Gen. Comp. Endocrinol. 2013, 190, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Lee, K.Y.; Han, J.Y. Precise genome editing in poultry and its application to industries. Genes 2020, 11, 1182. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, D.H.; Lee, K. Current approaches and applications in avian genome editing. Int. J. Mol. Sci. 2020, 21, 3937. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Park, T.S.; Lee, H.J.; Kim, K.H.; Kim, J.S.; Han, J.Y. Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. USA 2014, 111, 12716–12721. [Google Scholar] [CrossRef] [Green Version]
- Taylor, L.; Carlson, D.F.; Nandi, S.; Sherman, A.; Fahrenkrug, S.C.; McGrew, M.J. Efficient TALEN-mediated gene targeting of chicken primordial germ cells. Development 2017, 144, 928–934. [Google Scholar] [CrossRef] [Green Version]
- Ricroch, A. Global developments of genome editing in agriculture. Transgenic Res. 2019, 28, 45–52. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.H.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Bakhtiar, A.; Chowdhury, E.H. PH-responsive strontium nanoparticles for targeted gene therapy against mammary carcinoma cells. Asian J. Pharm. Sci. 2021, 16, 236–252. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Zhang, F.; Ding, Y. CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications. Pharmaceutics 2021, 13, 1649. [Google Scholar] [CrossRef]
- Branden, L.J.; Mohamed, A.J.; Smith, C.I.E. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999, 17, 784–787. [Google Scholar] [CrossRef]
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Yee, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, Y.; Onuma, H.; Sato, R.; Sato, Y.; Hashiba, A.; Maeki, M.; Tokeshi, M.; Kayesh, M.E.H.; Kohara, M.; Tsukiyama-Kohara, K.; et al. Lipid nanoparticles loaded with ribonucleoprotein-oligonucleotide complexes synthesized using a microfluidic device exhibit robust genome editing and hepatitis B virus inhibition. J. Control. Release 2021, 330, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.A. Pronuclear microinjection of one-cell embryos. Methods Mol. Biol. 2020, 2066, 27–33. [Google Scholar] [CrossRef]
- Salter, D.W.; Smith, E.J.; Hughes, S.H.; Wright, S.E.; Crittenden, L.B. Transgenic chickens: Insertion of retroviral genes into the chicken germ line. Virology 1987, 157, 236–240. [Google Scholar] [CrossRef]
- Mizuarai, S.; Ono, K.; Yamaguchi, K.; Nishijima, K.; Kamihira, M.; Iijima, S. Production of transgenic quails with high frequency of germ-line transmission using VSV-G pseudotyped retroviral vector. Biochem. Biophys. Res. Commun. 2001, 286, 456–463. [Google Scholar] [CrossRef]
- McGrew, M.J.; Sherman, A.; Ellard, F.M.; Lillico, S.G.; Gilhooley, H.J.; Kingsman, A.J.; Mitrophanous, K.A.; Sang, H. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 2004, 5, 728–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamihira, M.; Ono, K.; Esaka, K.; Nishijima, K.; Kigaku, R.; Komatsu, H.; Yamashita, T.; Kyogoku, K.; Iijima, S. High-level expression of single-chain Fv-Fc fusion protein in serum and egg white of genetically manipulated chickens by using a retroviral vector. J. Virol. 2005, 79, 10864–10874. [Google Scholar] [CrossRef] [Green Version]
- Chapman, S.C.; Lawson, A.; Macarthur, W.C.; Wiese, R.J.; Loechel, R.H.; Burgos-Trinidad, M.; Wakefield, J.K.; Ramabhadran, R.; Mauch, T.J.; Schoenwolf, G.C. Ubiquitous GFP expression in transgenic chicken using a lentiviral vector. Development 2005, 132, 935–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, B.C.; Kwon, M.S.; Choi, B.R.; Kim, J.H.; Cho, S.K.; Sohn, S.H.; Cho, E.J.; Lee, H.T.; Chang, W.; Jeon, I.; et al. Production of germline transgenic chickens expressing enhanced green fluorescent protein using a MoMLV-based retrovirus vector. FASEB J. 2006, 20, 2251–2260. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.B.; Lois, C. Generation of tissue-specific transgenic birds with lentiviral vectors. Proc. Natl. Acad. Sci. USA 2005, 102, 16443–16447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lillico, S.G.; Sherman, A.; McGrew, M.J.; Robertson, C.D.; Smith, J.; Haslam, C.; Barnard, P.; Radcliffe, P.A.; Mitrophanous, K.A.; Elliot, E.A.; et al. Oviduct-specific expression of two therapeutic proteins in transgenic hens. Proc. Natl. Acad. Sci. USA 2007, 104, 1771–1776. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Ma, J.; Lee, K. Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail. Proc. Natl. Acad. Sci. USA 2019, 116, 13288–13292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kim, D.H.; Lee, K. Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide. Int. J. Mol. Sci. 2020, 21, 1504. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Han, C.X.; Zhou, H.; Ding, J.M.; Xu, Z.; Yang, L.Y.; He, C.; Akinyemi, F.; Zheng, Y.M.; Qin, C.; et al. Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle. Int. J. Mol. Sci. 2020, 21, 2584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kim, D.-H.; Karolak, M.C.; Shin, S.; Lee, K. Generation of genome-edited chicken and duck lines by adenovirus-mediated in vivo genome editing. Proc. Natl. Acad. Sci. USA 2022, 119, e2214344119. [Google Scholar] [CrossRef]
- Adam, M.; Oualikene, W.; Le Cocq, H.; Guittet, M.; Eloit, M. Replication-defective adenovirus type 5 as an in vitro and in vivo gene transfer vector in chickens. J. Gen. Virol. 1995, 76, 3153–3157. [Google Scholar] [CrossRef]
- Shin, J.; Bae, D.R.; Latshaw, J.D.; Wick, M.P.; Reddish, J.M.; Lee, K. Technical note: A gene delivery system in the embryonic cells of avian species using a human adenoviral vector. J. Anim. Sci. 2009, 87, 2791–2795. [Google Scholar] [CrossRef] [Green Version]
- Pain, B.; Clark, M.E.; Shen, M.; Nakazawa, H.; Sakurai, M.; Samarut, J.; Etches, R.J. Long-term in vitro culture and characterisation of avian embryonic stem cells with multiple morphogenetic potentialities. Development 1996, 122, 2339–2348. [Google Scholar] [CrossRef]
- Park, T.S.; Hong, Y.H.; Kwon, S.C.; Lim, J.M.; Han, J.Y. Birth of germline chimeras by transfer of chicken embryonic germ (EG) cells into recipient embryos. Mol. Reprod. Dev. 2003, 65, 389–395. [Google Scholar] [CrossRef]
- Kim, J.N.; Park, T.S.; Park, S.H.; Park, K.J.; Kim, T.M.; Lee, S.K.; Lim, J.M.; Han, J.Y. Migration and proliferation of intact and genetically modified primordial germ cells and the generation of a transgenic chicken. Biol. Reprod. 2010, 82, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Swift, C.H. Origin and early history of the primordial germ cells in the chick. Am. J. Anat. 1914, 15, 483–516. [Google Scholar] [CrossRef] [Green Version]
- Tsunekawa, N.; Naito, M.; Sakai, Y.; Nishida, T.; Noce, T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development 2000, 127, 2741–2750. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Choi, H.J.; Lee, H.G.; Lim, J.M.; Ono, T.; Han, J.Y. DAZL expression explains origin and central formation of primordial germ cells in chickens. Stem Cells Dev. 2016, 25, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Murai, H.; Shibuya, M.; Kishita, R.; Sunase, C.; Tamura, K.; Saito, D. Envelopment by endothelial cells initiates translocation of avian primordial germ cell into vascular tissue. Dev. Dyn. 2021, 250, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- van de Lavoir, M.C.; Diamond, J.H.; Leighton, P.A.; Mather-Love, C.; Heyer, B.S.; Bradshaw, R.; Kerchner, A.; Hooi, L.T.; Gessaro, T.M.; Swanberg, S.E.; et al. Germline transmission of genetically modified primordial germ cells. Nature 2006, 441, 766–769. [Google Scholar] [CrossRef]
- Wentworth, B.C.; Tsai, H.; Hallett, J.H.; Gonzales, D.S.; Rajcic-Spasojevic, G. Manipulation of avian primordial germ cells and gonadal differentiation. Poult. Sci. 1989, 68, 999–1010. [Google Scholar] [CrossRef]
- Tajima, A.; Naito, M.; Yasuda, Y.; Kuwana, T. Production of germ line chimera by transfer of primordial germ cells in the domestic chicken (Gallus domesticus). Theriogenology 1993, 40, 509–519. [Google Scholar] [CrossRef]
- Chang, I.K.; Jeong, D.K.; Hong, Y.H.; Park, T.S.; Moon, Y.K.; Ohno, T.; Han, J.Y. Production of germline chimeric chickens by transfer of cultured primordial germ cells. Cell Biol. Int. 1997, 21, 495–499. [Google Scholar] [CrossRef]
- Park, T.S.; Jeong, D.K.; Kim, J.N.; Song, G.; Hong, Y.H.; Lim, J.M.; Han, J.Y. Improved germline transmission in chicken chimeras produced by transplantation of gonadal primordial germ cells into recipient embryos. Biol. Reprod. 2003, 68, 1657–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macdonald, J.; Taylor, L.; Sherman, A.; Kawakami, K.; Takahashi, Y.; Sang, H.M.; McGrew, M.J. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc. Natl. Acad. Sci. USA 2012, 109, 1466–1472. [Google Scholar] [CrossRef] [Green Version]
- Park, T.S.; Han, J.Y. A piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc. Natl. Acad. Sci. USA 2012, 109, 9337–9341. [Google Scholar] [CrossRef] [Green Version]
- Tyack, S.G.; Jenkins, K.A.; O’Neil, T.E.; Wise, T.G.; Morris, K.R.; Bruce, M.P.; McLeod, S.; Wade, A.J.; McKay, J.; Moore, R.J.; et al. A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells. Transgenic Res. 2013, 22, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; van de Lavoir, M.C.; Albanese, J.; Beenhouwer, D.O.; Cardarelli, P.M.; Cuison, S.; Deng, D.F.; Deshpande, S.; Diamond, J.H.; Green, L.; et al. Production of human monoclonal antibody in eggs of chimeric chickens. Nat. Biotechnol. 2005, 23, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, L.; Pedersen, D.; Ching, K.H.; Yi, H.; Collarini, E.J.; Izquierdo, S.; van de Lavoir, M.C.; Leighton, P.A. Germline Gene Editing in Chickens by Efficient CRISPR-Mediated Homologous Recombination in Primordial Germ Cells. PLoS ONE 2016, 11, e0154303. [Google Scholar] [CrossRef]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Tagami, T. Efficient production of human interferon β in the white of eggs from ovalbumin gene-targeted hens. Sci. Rep. 2018, 8, 10203. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Seo, M.; Choi, H.J.; Rengaraj, D.; Jung, K.M.; Park, J.S.; Lee, K.Y.; Kim, Y.M.; Park, K.J.; Han, S.T.; et al. DMRT1 gene disruption alone induces incomplete gonad feminization in chicken. FASEB J. 2021, 35, e21876. [Google Scholar] [CrossRef]
- Cooper, C.A.; Challagulla, A.; Jenkins, K.A.; Wise, T.G.; O’Neil, T.E.; Morris, K.R.; Tizard, M.L.; Doran, T.J. Generation of gene edited birds in one generation using sperm transfection assisted gene editing (STAGE). Transgenic Res. 2017, 26, 331–347. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.F.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Teng, F.; Li, T.D.; Zhou, Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, T.; Teng, F.; Guo, R.F.; Li, W.; Zhou, Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014, 24, 372–375. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Asami, M.; Perry, A.C. Asymmetric parental genome engineering by Cas9 during mouse meiotic exit. Sci. Rep. 2014, 4, 7621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, M.; Wu, Y.; Li, J. Generation and application of mammalian haploid embryonic stem cells. J. Intern. Med. 2016, 280, 236–245. [Google Scholar] [CrossRef] [Green Version]
- Mashiko, D.; Fujihara, Y.; Satouh, Y.; Miyata, H.; Isotani, A.; Ikawa, M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 2013, 3, 3355. [Google Scholar] [CrossRef] [Green Version]
- Hrabia, A.; Takagi, S.; Ono, T.; Shimada, K. Fertilization and development of quail oocytes after intracytoplasmic sperm injection. Biol. Reprod. 2003, 69, 1651–1657. [Google Scholar] [CrossRef]
- Naito, M.; Agata, K.; Otsuka, K.; Kino, K.; Ohta, M.; Hirose, K.; Perry, M.M.; Eguchi, G. Embryonic expression of ß-actin-lacZ hybrid gene injected into the fertilized ovum of the domestic fowl. Int. J. Dev. Biol. 1991, 35, 69–75. [Google Scholar] [PubMed]
- Ono, T. The complete in vitro development of quail embryo. In Transgenic Animals: Generation and Use; Houdebine, L.M., Ed.; Harwood Academic Publishers: Amsterdam, The Netherlands, 1997; Volume 3, pp. 61–67. [Google Scholar]
- Hrabia, A.; Takagi, S.; Shimada, K. Variable response to hormonal induction of multiple ovulation in quail. J. Poult. Sci. 2003, 40, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Ono, T.; Murakami, T.; Mochii, M.; Agata, K.; Kino, K.; Otsuka, K.; Ohta, M.; Mizutani, M.; Yoshida, M.; Eguchi, G. A complete culture system for avian transgenesis, supporting quail embryos from the single-cell stage to hatching. Dev. Biol. 1994, 161, 126–130. [Google Scholar] [CrossRef]
- Mizushima, S.; Takagi, S.; Ono, T.; Atsumi, Y.; Tsukada, A.; Saito, N.; Shimada, K. Developmental enhancement of intracytoplasmic sperm injection (ICSI)-generated quail embryos by phospholipase Cζ cRNA. J. Poult. Sci. 2008, 45, 152–158. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, S. Establishment of intracytoplasmic sperm injection technique in Japanese quail and its possible application for poultry resources and transgenic birds. J. Poult. Sci. 2012, 49, 225–230. [Google Scholar] [CrossRef] [Green Version]
- Harper, E.H. The fertilization and early development of the pigeon’s egg. Am. J. Anat. 1904, 3, 349–386. [Google Scholar] [CrossRef] [Green Version]
- Patterson, J.T. Studies on the early development of the hen’s egg. I. History of the early cleavage and of the accessory cleavage. J. Morphol. 1910, 21, 101–134. [Google Scholar] [CrossRef] [Green Version]
- Fofanova, K.A. Morphologic data on polyspermy in chickens. Fed. Proc. Transl. Suppl. 1965, 24, 239–247. [Google Scholar]
- Nakanishi, A.; Utsumi, K.; Iritani, A. Early nuclear events of in vitro fertilization in the domestic fowl (Gallus domesticus). Mol. Reprod. Dev. 1990, 26, 217–221. [Google Scholar] [CrossRef]
- Elinson, R.P. Fertilization in amphibians: The ancestry of the block to polyspermy. Int. Rev. Cytol. 1986, 101, 59–100. [Google Scholar] [CrossRef]
- Wong, J.L.; Wessel, G.M. Defending the zygote: Search for the ancestral animal block to polyspermy. Curr. Top Dev. Biol. 2006, 72, 1–151. [Google Scholar] [CrossRef]
- Iwao, Y. Egg activation in physiological polyspermy. Reproduction 2012, 144, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Eyal-Giladi, H.; Kochav, S. From cleavage to primitive streak formation: A complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev. Biol. 1976, 49, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Perry, M.M. Nuclear events from fertilization to the early cleavage stages in the domestic fowl (Gallus domesticus). J. Anat. 1987, 150, 99–109. [Google Scholar] [PubMed]
- Stepińska, U.; Olszańska, B. Detection of deoxyribonuclease I and II activities in Japanese quail oocytes. Zygote 2001, 9, 1–7. [Google Scholar] [CrossRef]
- Stepińska, U.; Olszańska, B. DNase I and II present in avian oocytes: A possible involvement in sperm degradation at polyspermic fertilisation. Zygote 2003, 11, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Stepińska, U.; Olszańska, B. Molecular aspects of avian oogenesis and fertilisation. Int. J. Dev. Biol. 2008, 52, 187–194. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, S. Fertilization 2: Polyspermic fertilization. Adv. Exp. Med. Biol. 2017, 1001, 105–123. [Google Scholar] [CrossRef]
- Stricker, S.A. Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev. Biol. 1999, 211, 157–176. [Google Scholar] [CrossRef]
- Runft, L.L.; Jaffe, L.A.; Mehlmann, L.M. Egg activation at fertilization: Where it all begins. Dev. Biol. 2002, 245, 237–254. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Yoshida, N.; Suzuki, E.; Okuda, E.; Perry, A.C. Full-term mouse development by abolishing Zn2+-dependent metaphase II arrest without Ca2+ release. Development 2010, 137, 2659–2669. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, S.; Takagi, S.; Ono, T.; Atsumi, Y.; Tsukada, A.; Saito, N.; Shimada, K. Possible role of calcium on oocyte development after intracytoplasmic sperm injection in quail (Coturnix japonica). J. Exp. Zool. 2007, 307A, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, S.; Hiyama, G.; Shiba, K.; Inaba, K.; Dohra, H.; Ono, T.; Shimada, K.; Sasanami, T. The birth of quail chicks after intracytoplasmic sperm injection. Development 2014, 141, 3799–3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, C.M.; Larman, M.G.; Parrington, J.; Cox, L.J.; Royse, J.; Blayney, L.M.; Swann, K.; Lai, F.A. PLC zeta: A sperm-specific trigger of Ca2+ oscillations in eggs and embryo development. Development 2002, 129, 3533–3544. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Matsumoto, T.; Hirahara, S.; Nakashima, A.; Ueno, S.; Oda, S.; Miyazaki, S.; Iwao, Y. Characterization of a sperm factor for egg activation at fertilization of the newt Cynops pyrrhogaster. Dev. Biol. 2007, 306, 797–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, S.; Takagi, S.; Ono, T.; Atsumi, Y.; Tsukada, A.; Saito, N.; Shimada, K. Phospholipase Cζ mRNA expression and its potency during spermatogenesis for activation of quail oocyte as a sperm factor. Mol. Reprod. Dev. 2009, 76, 1200–1207. [Google Scholar] [CrossRef]
- Mizushima, S.; Matsuzaki, M.; Sasanami, T. Handling of gametes for in vitro insemination in birds. Methods Mol. Biol. 2017, 1650, 243–257. [Google Scholar] [CrossRef]
- Love, J.; Gribbin, C.; Mather, C.; Sang, H. Transgenic birds by DNA microinjection. Biotechnology 1994, 12, 60–63. [Google Scholar] [CrossRef]
- Naito, M.; Sasaki, E.; Ohtaki, M.; Sakurai, M. Introduction of exogenous DNA into somatic and germ cells of chickens by microinjection into the germinal disc of fertilized ova. Mol. Reprod. Dev. 1994, 37, 167–171. [Google Scholar] [CrossRef]
- Sherman, A.; Dawson, A.; Mather, C.; Gilhooley, H.; Li, Y.; Mitchell, R.; Finnegan, D.; Sang, H. Transposition of the Drosophila element mariner into the chicken germ line. Nat. Biotechnol. 1998, 16, 1050–1053. [Google Scholar] [CrossRef]
- Mizushima, S.; Sasanami, T.; Ono, T.; Matsuzaki, M.; Kansaku, N.; Kuroiwa, A. Cyclin D1 gene expression is essential for cell cycle progression from the maternal-to zygotic transition during blastoderm development in Japanese quail. Dev. Biol. 2021, 476, 249–258. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mizushima, S.; Sasanami, T.; Ono, T.; Kuroiwa, A. Current Approaches to and the Application of Intracytoplasmic Sperm Injection (ICSI) for Avian Genome Editing. Genes 2023, 14, 757. https://doi.org/10.3390/genes14030757
Mizushima S, Sasanami T, Ono T, Kuroiwa A. Current Approaches to and the Application of Intracytoplasmic Sperm Injection (ICSI) for Avian Genome Editing. Genes. 2023; 14(3):757. https://doi.org/10.3390/genes14030757
Chicago/Turabian StyleMizushima, Shusei, Tomohiro Sasanami, Tamao Ono, and Asato Kuroiwa. 2023. "Current Approaches to and the Application of Intracytoplasmic Sperm Injection (ICSI) for Avian Genome Editing" Genes 14, no. 3: 757. https://doi.org/10.3390/genes14030757