Terrain Ruggedness and Canopy Height Predict Short-Range Dispersal in the Critically Endangered Black-and-White Ruffed Lemur
Abstract
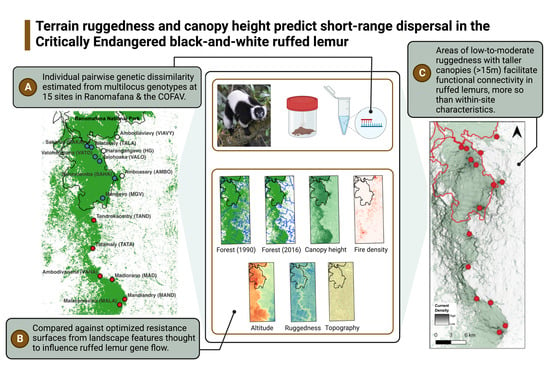
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Genetic Sampling, Relatedness, and Differentiation
2.3. Landcover Classification and Landscape Feature Selection
2.4. Resistance Surface Parameterization and Optimization
2.5. Resistance Surface Model Selection
2.6. Gravity Model
3. Results
3.1. Resistance Analysis
3.2. Gravity Model
4. Discussion
4.1. Between-Site Influences on Ruffed Lemur Dispersal
4.2. Within-Site Influences on Ruffed Lemur Dispersal
4.3. Spatial Variation in Response to Environmental Variables
4.4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nathan, R.; Getz, W.M.; Revilla, E.; Holyoak, M.; Kadmon, R.; Saltz, D.; Smouse, P.E. A movement ecology paradigm for unifying organismal movement research. Proc. Natl. Acad. Sci. USA 2008, 105, 19052–19059. [Google Scholar] [CrossRef] [Green Version]
- Neumann, W.; Martinuzzi, S.; Estes, A.B.; Pidgeon, A.M.; Dettki, H.; Ericsson, G.; Radeloff, V.C. Opportunities for the application of advanced remotely-sensed data in ecological studies of terrestrial animal movement. Mov. Ecol. 2015, 3, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaudrot, L.H.; Marshall, A.J. Primate communities are structured more by dispersal limitation than by niches. J. Anim. Ecol. 2011, 80, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Matthysen, E. Multicausality of dispersal: A review. In Dispersal Ecology and Evolution; Oxford University Press: Oxford, UK, 2012. [Google Scholar] [CrossRef]
- Schilthuizen, M. Ecotone: Speciation-prone. Trends Ecol. Evol. 2000, 15, 130–131. [Google Scholar] [CrossRef]
- Seehausen, O. African cichlid fish: A model system in adaptive radiation research. Proc. R. Soc. B Biol. Sci. 2006, 273, 1987–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, A.J.; Beaudrot, L.; Wittmer, H.U. Responses of primates and other frugivorous vertebrates to plant resource variability over space and time at Gunung Palung National Park. Int. J. Primatol. 2014, 35, 1178–1201. [Google Scholar] [CrossRef]
- Holyoak, M.; Casagrandi, R.; Nathan, R.; Revilla, E.; Spiegel, O. Trends and missing parts in the study of movement ecology. Proc. Natl. Acad. Sci. USA 2008, 105, 19060–19065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricketts, T.H. The matrix matters: Effective isolation in fragmented landscapes. Am. Nat. 2001, 158, 87–99. [Google Scholar] [CrossRef]
- Van Oort, H.; McLellan, B.N.; Serrouya, R. Fragmentation, dispersal and metapopulation function in remnant populations of endangered mountain caribou. Anim. Conserv. 2011, 14, 215–224. [Google Scholar] [CrossRef]
- Eycott, A.; Watts, K.; Brandt, G.; Buyung-Ali, L.; Bowler, D.; Stewart, G.; Pullin, A. Do landscape matrix features affect species movement? Collab. Environ. Evid. 2010, 8, 1–119. [Google Scholar]
- Eycott, A.E.; Stewart, G.B.; Buyung-Ali, L.M.; Bowler, D.E.; Watts, K.; Pullin, A.S. A meta-analysis on the impact of different matrix structures on species movement rates. Landsc. Ecol. 2012, 27, 1263–1278. [Google Scholar] [CrossRef]
- Bowler, D.E.; Benton, T.G. Variation in dispersal mortality and dispersal propensity among individuals: The effects of age, sex and resource availability. J. Anim. Ecol. 2009, 78, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.A.; Fryxell, J.M.; Thompson, I.D.; Baker, J.A. Mortality risk increases with natal dispersal distance in American martens. Proc. R. Soc. B Biol. Sci. 2009, 276, 3361–3367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowicki, P.; Vrabec, V.; Binzenhöfer, B.; Feil, J.; Zakšek, B.; Hovestadt, T.; Settele, J. Butterfly dispersal in inhospitable matrix: Rare, risky, but long-distance. Landsc. Ecol. 2014, 29, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Ries, L.; Debinski, D.M. Butterfly responses to habitat edges in the highly fragmented prairies of Central Iowa. J. Anim. Ecol. 2001, 70, 840–852. [Google Scholar] [CrossRef] [Green Version]
- Schtickzelle, N.; Baguette, M. Behavioural responses to habitat patch boundaries restrict dispersal and generate emigration–patch area relationships in fragmented landscapes. J. Anim. Ecol. 2003, 72, 533–545. [Google Scholar] [CrossRef]
- Smith, J.E.; Batzli, G.O. Dispersal and mortality of prairie voles (Microtus ochrogaster) in fragmented landscapes: A field experiment. Oikos 2006, 112, 209–217. [Google Scholar] [CrossRef]
- Baguette, M.; Legrand, D.; Fréville, H.; Van Dyck, H.; Ducatez, S. Evolutionary ecology of dispersal in fragmented landscape. In Dispersal Ecology and Evolution; Oxford University Press: Oxford, UK, 2012. [Google Scholar] [CrossRef]
- Pflüger, F.J.; Balkenhol, N. A plea for simultaneously considering matrix quality and local environmental conditions when analysing landscape impacts on effective dispersal. Mol. Ecol. 2014, 23, 2146–2156. [Google Scholar] [CrossRef] [Green Version]
- Gursky, S. Dispersal patterns in Tarsius spectrum. Int. J. Primatol. 2010, 31, 117–131. [Google Scholar] [CrossRef]
- Lin, Y.-T.K.; Batzli, G.O. The influence of habitat quality on dispersal, demography, and population dynamics of voles. Ecol. Monogr. 2001, 71, 245. [Google Scholar] [CrossRef]
- Lurz, P.W.W.; Garson, P.J.; Wauters, L.A. Effects of temporal and spatial variation in habitat quality on red squirrel dispersal behaviour. Anim. Behav. 1997, 54, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayor, L.S. Effects of habitat quality on growth, age of first reproduction, and dispersal in Gunnison’s prairie dogs (Cynomys gunnisoni). Can. J. Zool. 1985, 63, 2835–2840. [Google Scholar] [CrossRef]
- Sedgwick, J.A. Site fidelity, territory fidelity, and natal philopatry in Willow Flycatchers (Empidonax traillii). Auk 2004, 121, 1103–1121. [Google Scholar] [CrossRef]
- Stacey, P.B.; Ligon, J.D. Territory quality and dispersal options in the acorn woodpecker, and a challenge to the habitat-saturation model of cooperative breeding. Am. Nat. 1987, 130, 654–676. [Google Scholar] [CrossRef]
- Sutherland, G.D.; Harestad, A.S.; Price, K.; Lertzman, K. Scaling of natal dispersal distances in terrestrial birds and mammals. Ecol. Soc. 2000, 4, 16. [Google Scholar] [CrossRef]
- Whitmee, S.; Orme, C.D.L. Predicting dispersal distance in mammals: A trait-based approach. J. Anim. Ecol. 2013, 82, 211–221. [Google Scholar] [CrossRef]
- Ciucci, P.; Reggioni, W.; Maiorano, L.; Boitani, L. Long-distance dispersal of a rescued wolf from the northern Apennines to the western Alps. J. Wildl. Manag. 2009, 73, 1300–1306. [Google Scholar] [CrossRef]
- Fattebert, J.; Dickerson, T.; Balme, G.; Slotow, R.; Hunter, L. Long-distance natal dispersal in leopard reveals potential for a three-country metapopulation. S. Afr. J. Wildl. Res. 2013, 43, 61–67. [Google Scholar] [CrossRef]
- Krishnamurthy, R.; Cushman, S.A.; Sarkar, M.S.; Malviya, M.; Naveen, M.; Johnson, J.A.; Sen, S. Multi-scale prediction of landscape resistance for tiger dispersal in central India. Landsc. Ecol. 2016, 31, 1355–1368. [Google Scholar] [CrossRef]
- Slater, K.; Barrett, A.; Brown, L.R. Home range utilization by chacma baboon (Papio ursinus) troops on Suikerbosrand Nature Reserve, South Africa. PLoS ONE 2018, 13, e0194717. [Google Scholar] [CrossRef] [Green Version]
- Casillas, S.; Barbadilla, A. Molecular population genetics. Genetics 2017, 205, 1003–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tischendorf, L.; Fahrig, L. On the usage and measurement of landscape connectivity. Oikos 2000, 90, 7–19. [Google Scholar] [CrossRef] [Green Version]
- Manel, S.; Holderegger, R. Ten years of landscape genetics. Trends Ecol. Evol. 2013, 28, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Murphy, M. Package ‘GeNetIt.’ R Package Version 0.1-3. 2020. Available online: https://mran.microsoft.com/snapshot/2017-01-31/web/packages/GeNetIt/citation.html (accessed on 11 January 2021).
- Westphal, D.; Mancini, A.N.; Baden, A.L. Primate landscape genetics: A review and practical guide. Evol. Anthropol. Issues News Rev. 2021, 30, 171–184. [Google Scholar] [CrossRef]
- Goossens, B.; Chikhi, L.; Jalil, M.F.; Ancrenaz, M.; Lackman-Ancrenaz, I.; Mohamed, M.; Andau, P.; Bruford, M.W. Patterns of genetic diversity and migration in increasingly fragmented and declining orang-utan (Pongo pygmaeus) populations from Sabah, Malaysia. Mol. Ecol. 2005, 14, 441–456. [Google Scholar] [CrossRef]
- Quéméré, E.; Crouau-Roy, B.; Rabarivola, C.; Louis, E.E.; Chikhi, L. Landscape genetics of an endangered lemur (Propithecus tattersalli) within its entire fragmented range. Mol. Ecol. 2010, 19, 1606–1621. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, B.; Wu, R.; Zhao, L.; Hao, Y.; Wang, B.; Wei, F.; Long, Y.; Li, M. The effect of landscape features on population genetic structure in Yunnan snub-nosed monkeys (Rhinopithecus bieti) implies an anthropogenic genetic discontinuity. Mol. Ecol. 2009, 18, 3831–3846. [Google Scholar] [CrossRef]
- Blair, M.E.; Melnick, D.J. Scale-dependent effects of a heterogeneous landscape on genetic differentiation in the Central American squirrel monkey (Saimiri oerstedii). PLoS ONE 2012, 7, e43027. [Google Scholar] [CrossRef]
- Li, L.; Xue, Y.; Wu, G.; Li, D.; Giraudoux, P. Potential habitat corridors and restoration areas for the black-and-white snub-nosed monkey Rhinopithecus bieti in Yunnan, China. Oryx 2014, 49, 719–726. [Google Scholar] [CrossRef] [Green Version]
- Moraes, A.M.; Ruiz-Miranda, C.R.; Galetti, P.M., Jr.; Niebuhr, B.B.; Alexandre, B.R.; Muylaert, R.L.; Grativol, A.D.; Ribeiro, J.W.; Ferreira, A.N.; Ribeiro, M.C. Landscape resistance influences effective dispersal of endangered golden lion tamarins within the Atlantic Forest. Biol. Conserv. 2018, 224, 178–187. [Google Scholar] [CrossRef] [Green Version]
- Oklander, L.I.; Miño, C.I.; Fernández, G.; Caputo, M.; Corach, D. Genetic structure in the southernmost populations of black-and-gold howler monkeys (Alouatta caraya) and its conservation implications. PLoS ONE 2017, 12, e0185867. [Google Scholar] [CrossRef] [Green Version]
- Baden, A.; Mancini, A.; Federman, S.; Holmes, S.M.; Johnson, S.E.; Kamilar, J.; Louis, E.E.; Bradley, B.J. Anthropogenic pressures drive population genetic structuring across a Critically Endangered lemur species range. Nat. Sci. Rep. 2019, 9, 16276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Lopez, M.J.; Barelli, C.; Rovero, F.; Hodges, K.; Roos, C.; Peterman, W.E.; Ting, N. A novel landscape genetic approach demonstrates the effects of human disturbance on the Udzungwa red colobus monkey (Procolobus gordonorum). Heredity 2016, 116, 167–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aylward, C.M.; Murdoch, J.D.; Kilpatrick, C.W. Multiscale landscape genetics of American marten at their southern range periphery. Heredity 2020, 124, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Balkenhol, N.; Schwartz, M.K.; Inman, R.M.; Copeland, J.P.; Squires, J.S.; Anderson, N.J.; Waits, L.P. Landscape genetics of wolverines (Gulo gulo): Scale-dependent effects of bioclimatic, topographic, and anthropogenic variables. J. Mammal. 2020, 101, 790–803. [Google Scholar] [CrossRef]
- Kierepka, E.M.; Anderson, S.J.; Swihart, R.K.; Rhodes, O.E. Differing, multiscale landscape effects on genetic diversity and differentiation in eastern chipmunks. Heredity 2020, 124, 457–468. [Google Scholar] [CrossRef]
- Millette, K.L.; Keyghobadi, N. The relative influence of habitat amount and configuration on genetic structure across multiple spatial scales. Ecol. Evol. 2015, 5, 73–86. [Google Scholar] [CrossRef]
- Koen, E.L.; Bowman, J.; Wilson, P.J. Node-based measures of connectivity in genetic networks. Mol. Ecol. Resour. 2016, 16, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Balko, E.A.; Underwood, B.H. Effects of forest structure and composition on food availability for Varecia variegata at Ranomafana National Park, Madagascar. Am. J. Primatol. 2005, 66, 45–70. [Google Scholar] [CrossRef]
- Herrera, J.P.; Wright, P.C.; Lauterbur, E.; Ratovonjanahary, L.; Taylor, L.L. The Effects of habitat disturbance on lemurs at Ranomafana National Park, Madagascar. Int. J. Primatol. 2011, 32, 1091–1108. [Google Scholar] [CrossRef]
- Morelli, T.L.; Smith, A.B.; Mancini, A.N.; Balko, E.A.; Borgerson, C.; Dolch, R.; Farris, Z.; Federman, S.; Golden, C.D.; Holmes, S.M.; et al. The fate of Madagascar’s rainforest habitat. Nat. Clim. Chang. 2020, 10, 89–96. [Google Scholar] [CrossRef]
- Ratsimbazafy, H.J. On the Brink of Extinction and the Process of Recovery: Responses of Black-And-White Rufffed Lemurs (Varecia Variegata Variegata) to Disturbance in Manombo Forest, Madagascar; Stony Brook University: Stony Brook, NY, USA, 2002. [Google Scholar]
- White, F.J.; Overdorff, D.J.; Balko, E.A.; Wright, P.C. Distribution of ruffed lemurs (Varecia variegata) in Ranomafana National Park, Madagascar. Folia Primatol. 1995, 64, 124–131. [Google Scholar] [CrossRef]
- Baden, A.L.; Holmes, S.M.; Johnson, S.E.; Engberg, S.E.; Louis, E.E.; Bradley, B.J. Species-level view of population structure and gene flow for a Critically Endangered primate (Varecia variegata). Ecol. Evol. 2014, 4, 2675–2692. [Google Scholar] [CrossRef]
- Mancini, A.N. Environmental Drivers of Dispersal in Black-and-White Ruffed Lemurs (Varecia Variegata); Graduate Center of the City University of New York: New York, NY, USA, 2023. [Google Scholar]
- Barrett, M.A.; Brown, J.L.; Morikawa, M.K.; Labat, J.-N.; Yoder, A.D. CITES Designation for Endangered Rosewood in Madagascar. Science 2010, 328, 1109–1110. [Google Scholar] [CrossRef]
- Cabeza, M.; Terraube, J.; Burgas, D.; Temba, E.M.; Rakoarijaoana, M. Gold is not green: Artisanal gold mining threatens Ranomafana National Park’s biodiversity. Anim. Conserv. 2019, 22, 417–419. [Google Scholar] [CrossRef]
- Van Vliet, N.; Mertz, O.; Heinimann, A.; Langanke, T.; Pascual, U.; Schmook, B.; Adams, C.; Schmidt-Vogt, D.; Messerli, P.; Leisz, S.; et al. Trends, drivers and impacts of changes in swidden cultivation in tropical forest-agriculture frontiers: A global assessment. Glob. Environ. Chang. 2012, 22, 418–429. [Google Scholar] [CrossRef]
- Beeby, N.; Baden, A.L. Seasonal variability in the diet and feeding ecology of black-and-white ruffed lemurs (Varecia variegata) in Ranomafana National Park, southeastern Madagascar. Am. J. Phys. Anthropol. 2021, 174, 763–775. [Google Scholar] [CrossRef]
- Epps, C.; Keyghobadi, N. Landscape genetics in a changing world: Disentangling historical and contemporary influences and inferring change. Mol. Ecol. 2015, 24, 6021–6040. [Google Scholar] [CrossRef] [Green Version]
- Aiba, S.; Kitayama, K. Structure, composition and species diversity in an altitude-substrate matrix of rain forest tree communities on Mount Kinabalu, Borneo. Plant Ecol. 1999, 140, 139–157. [Google Scholar] [CrossRef]
- Asner, G.P.; Anderson, C.B.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Sinca, F.; Malhi, Y. Landscape-scale changes in forest structure and functional traits along an Andes-to-Amazon elevation gradient. Biogeosciences 2014, 11, 843–856. [Google Scholar] [CrossRef] [Green Version]
- Lieberman, D.; Lieberman, M.; Peralta, R.; Hartshorn, G.S. Tropical forest structure and composition on a large-scale altitudinal gradient in Costa Rica. J. Ecol. 1996, 84, 137–152. [Google Scholar] [CrossRef]
- Homeier, J.; Breckle, S.-W.; Günter, S.; Rollenbeck, R.T.; Leuschner, C. Tree diversity, forest structure and productivity along altitudinal and topographical gradients in a species-rich Ecuadorian montane rain forest. Biotropica 2010, 42, 140–148. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sagar, R.; Singh, J.S. Tropical forest structure and diversity in relation to altitude and disturbance in a Biosphere Reserve in central India. Appl. Veg. Sci. 2008, 11, 461–470. [Google Scholar] [CrossRef]
- Sharma, C.M.; Suyal, S.; Gairola, S.; Ghildiyal, S.K. Tree species composition and diversity along an altitudinal gradient in moist tropical montane valley slopes of the Garhwal Himalaya, India. For. Sci. Technol. 2009, 5, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Whittaker, R.H.; Niering, W.A. Vegetation of the Santa Catalina Mountains, Arizona. V. Biomass 1975, production, and diversity along the elevation gradient. Ecology 1975, 56, 771–790. [Google Scholar] [CrossRef] [Green Version]
- Kalinowski, S. User’s manual for ML-Relate input files null alleles. Mol. Ecol. Notes 2006, 6, 576–579. [Google Scholar] [CrossRef]
- Milligan, B.G. Maximum-likelihood estimation of relatedness. Genetics 2003, 1167, 1153–1167. [Google Scholar] [CrossRef]
- Rodríguez-Ramila, S.T.; Wang, J. The effect of close relatives on unsupervised Bayesian clustering algorithms in population genetic structure analysis. Mol. Ecol. Resour. 2012, 12, 873–884. [Google Scholar] [CrossRef]
- Peterman, W.; Brocato, E.R.; Semlitsch, R.D.; Eggert, L.S. Reducing bias in population and landscape genetic inferences: The effects of sampling related individuals and multiple life stages. PeerJ 2016, 4, e1813. [Google Scholar] [CrossRef] [Green Version]
- Waples, R.S.; Anderson, E.C. Purging putative siblings from population genetic data sets: A cautionary view. Mol. Ecol. 2017, 26, 1211–1224. [Google Scholar] [CrossRef] [Green Version]
- Rousset, F. Genetic differentiation between individuals. J. Evol. Biol. 2000, 13, 58–62. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, A Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3; Lausanne University: Lausanne, Switzerland, 2001. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 11 January 2021).
- Gerber, B.D. Madagascar GIS: Spatial information to Aid Research and Conservation. 2010. Available online: http://filebox.vt.edu/users/bgerber/services.htm (accessed on 2 April 2016).
- Hijmans, R.J.; Guarino, L.; Bussink, C.; Mathur, P.; Cruz, M.; Barrentes, I.; Rojas, E.; DIVA-GIS. A Geographic Information System for the Analysis of Species Distribution Data. 2004. Available online: http://www.diva-gis.org (accessed on 5 January 2021).
- Riley, S.J.; DeGloria, S.D.; Elliot, R. A terrain ruggedness index that quantifies topographic heterogeneity. Intermt. J. Sci. 1999, 5, 23–27. [Google Scholar]
- Giglio, L.; Schroeder, W.; Hall, J.V.; Justice, C.O. MODIS Collection 4 Active Fire Product User’s Guide Table of Contents. Revision B; Nasa: Washington, DC, USA, 2018; Available online: https://modis-fire.umd.edu/files/MODIS_C6_Fire_User_Guide_B.pdf (accessed on 4 January 2021).
- Page, W.G.; Wagenbrenner, N.S.; Butler, B.W.; Blunck, D.L. An analysis of spotting distances during the 2017 fire season in the Northern Rockies, USA. Can. J. For. Res. 2019, 49, 317–325. [Google Scholar] [CrossRef]
- Potapov, P.; Li, X.; Hernandez-Serna, A.; Tyukavina, A.; Hansen, M.C.; Kommareddy, A.; Pickens, A.; Turubanova, S.; Tang, H.; Silva, C.E.; et al. Mapping global forest canopy height through integration of GEDI and Landsat data. Remote Sens. Environ. 2021, 253, 112165. [Google Scholar] [CrossRef]
- Cushman, S.A.; Landguth, E.L. Scale dependent inference in landscape genetics. Landsc. Ecol. 2010, 25, 967–979. [Google Scholar] [CrossRef]
- McRae, B.H.; Beier, P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. USA 2007, 104, 19885–19890. [Google Scholar] [CrossRef] [Green Version]
- Van Etten, J. R package gdistance: Distances and routes on geographical grids. J. Stat. Softw. 2017, 76, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Peterman, B. A Vignette/Tutorial to Use ResistanceGA. 2017. Available online: https://petermanresearch.weebly.com/uploads/2/5/9/2/25926970/resistancega.pdf (accessed on 21 December 2021).
- Bolliger, J.; Lander, T.; Balkenhol, N. Landscape genetics since 2003: Status, challenges and future directions. Landsc. Ecol. 2014, 29, 361–366. [Google Scholar] [CrossRef] [Green Version]
- Zeller, K.A.; McGarigal, K.; Whiteley, A.R. Estimating landscape resistance to movement: A review. Landsc. Ecol. 2012, 27, 777–797. [Google Scholar] [CrossRef]
- Towns, J.; Cockerill, T.; Dahan, M.; Foster, I.; Gaither, K.; Grimshaw, A.; Hazlewood, V.; Lathrop, S.; Lifka, D.; Peterson, G.D.; et al. XSEDE: Accelerating Scientific Discovery. Comput. Sci. Eng. 2014, 16, 62–74. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Van Strien, M.J.; Keller, D.; Holderegger, R. A new analytical approach to landscape genetic modelling: Least-cost transect analysis and linear mixed models. Mol. Ecol. 2012, 21, 4010–4023. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. lme4: Linear mixed-effects models using Eigen and S4. J. Stat. Softw. 2014, 67, 1–127. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Peterman, W.E. ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 2018, 9, 1638–1647. [Google Scholar] [CrossRef] [Green Version]
- Fotheringham, A.S.; O’Kelly, M.E. Spatial Interaction Models: Formulation and Applications; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1989. [Google Scholar]
- Murphy, M.A.; Dezzani, R.; Pilliod, D.S.; Storfer, A. Landscape genetics of high mountain frog metapopulations. Mol. Ecol. 2010, 19, 3634–3649. [Google Scholar] [CrossRef]
- Robertson, J.M.; Murphy, M.A.; Pearl, C.A.; Adams, M.J.; Páez-Vacas, M.I.; Haig, S.M.; Pilliod, D.S.; Storfer, A.; Funk, W.C. Regional variation in drivers of connectivity for two frog species (Rana pretiosa and R. luteiventris) from the U.S. Pacific Northwest. Mol. Ecol. 2018, 27, 3242–3256. [Google Scholar] [CrossRef]
- Wishingrad, V.; Thomson, R.C. Testing concordance and conflict in spatial replication of landscape genetics inferences. bioRxiv 2021, bioRxiv:2021.06.21.449301. [Google Scholar] [CrossRef]
- Vergara, M.; Cushman, S.A.; Ruiz-González, A. Ecological differences and limiting factors in different regional contexts: Landscape genetics of the stone marten in the Iberian Peninsula. Landsc. Ecol. 2017, 32, 1269–1283. [Google Scholar] [CrossRef]
- Hohnen, R.; Tuft, K.D.; Legge, S.; Hillyer, M.; Spencer, P.B.; Radford, I.J.; Johnson, C.N.; Burridge, C.P. Rainfall and topography predict gene flow among populations of the declining northern quoll (Dasyurus hallucatus). Conserv. Genet. 2016, 17, 1213–1228. [Google Scholar] [CrossRef]
- Dröge, S.; Poudyal, M.; Hockley, N.; Mandimbiniaina, R.; Rasoamanana, A.; Andrianantenaina, N.S.; Llopis, J.C. Constraints on rice cultivation in eastern Madagascar: Which factors matter to smallholders, and which influence food security? Hum. Ecol. 2022, 50, 493–513. [Google Scholar] [CrossRef]
- Whitman, K.; Ratsimbazafy, J.; Stevens, N. The use of System of Rice Intensification (SRI) near Maromizaha Protected Area, Madagascar. Madag. Conserv. Dev. 2020, 15, 1–8. [Google Scholar]
- Milanesi, P.; Holderegger, R.; Bollmann, K.; Gugerli, F.; Zellweger, F. Three-dimensional habitat structure and landscape genetics: A step forward in estimating functional connectivity. Ecology 2017, 98, 393–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borja-Martínez, G.; Tapia-Flores, D.; Shafer, A.B.A.; Vázquez-Domínguez, E. Highland forest’s environmental complexity drives landscape genomics and connectivity of the rodent Peromyscus melanotis. Landsc. Ecol. 2022, 37, 1653–1671. [Google Scholar] [CrossRef]
- Olah, G.; Smith, A.L.; Asner, G.P.; Brightsmith, D.J.; Heinsohn, R.G.; Peakall, R. Exploring dispersal barriers using landscape genetic resistance modelling in scarlet macaws of the Peruvian Amazon. Landsc. Ecol. 2017, 32, 445–456. [Google Scholar] [CrossRef]
- Baden, A.L. Communal Infant Care in Black-and-White Ruffed Lemurs (Varecia Variegata); Stony Brook University: New York, NY, USA, 2011. [Google Scholar]
- Balko, E.A. A Behaviorally Plastic Response to Forest Composition and Logging Disturbance by Varecia Variegata Variegata in Ranomafana National Park, Madagascar; State University of New York: New York, NY, USA, 1998. [Google Scholar]
- Goodman, S.M.; Ganzhorn, J.U. Biogeography of lemurs in the humid forests of Madagascar: The role of elevational distribution and rivers. J. Biogeogr. 2004, 31, 47–55. [Google Scholar] [CrossRef]
- Lehman, S.M.; Ratsimbazafy, J.; Rajaonson, A.; Day, S. Decline of Propithecus diadema edwardsi and Varecia variegata variegata (Primates: Lemuridae) in south-east Madagascar. Oryx 2006, 40, 108–111. [Google Scholar] [CrossRef] [Green Version]
- Hauser, S.S.; Leberg, P.L. Riparian areas potentially provide crucial corridors through fragmented landscape for black-capped vireo (Vireo atricapilla) source-sink system. Conserv. Genet. 2021, 22, 1–10. [Google Scholar] [CrossRef]
- Parsley, M.B.; Torres, M.L.; Banerjee, S.M.; Tobias, Z.J.C.; Goldberg, C.S.; Murphy, M.A.; Mims, M.C. Multiple lines of genetic inquiry reveal effects of local and landscape factors on an amphibian metapopulation. Landsc. Ecol. 2020, 35, 319–335. [Google Scholar] [CrossRef]
- Hodson, J.; Fortin, D.; Bélanger, L.; Journal, S.; June, N.; Hodson, J.; Bélanger, L. Fine-scale disturbances shape space-use patterns of a boreal forest herbivore. J. Mammal. 2010, 91, 607–619. [Google Scholar] [CrossRef] [Green Version]
- McLean, K.A.; Jansen, P.A.; Crofoot, M.C.; Asner, G.P.; Hopkins, M.E.; Martin, R.E.; Knapp, D.E. Movement patterns of three arboreal primates in a Neotropical moist forest explained by LiDAR-estimated canopy structure. Landsc. Ecol. 2016, 31, 1849–1862. [Google Scholar] [CrossRef] [Green Version]
- Moriarty, K.M.; Epps, C.W.; Betts, M.G.; Hance, D.J.; Bailey, J.D.; Zielinski, W.J. Experimental evidence that simplified forest structure interacts with snow cover to influence functional connectivity for Pacific martens. Landsc. Ecol. 2015, 30, 1865–1877. [Google Scholar] [CrossRef]
- Moriarty, K.M.; Epps, C.W.; Zielinski, W.J. Forest thinning changes movement patterns and habitat use by Pacific marten. J. Wildl. Manag. 2016, 80, 621–633. [Google Scholar] [CrossRef]
- Palminteri, S.; Powell, G.V.N.; Asner, G.P.; Peres, C.A. LiDAR measurements of canopy structure predict spatial distribution of a tropical mature forest primate. Remote Sens. Environ. 2012, 127, 98–105. [Google Scholar] [CrossRef]
- Wenner, S.M.; Murphy, M.A.; Delaney, K.S.; Pauly, G.B.; Richmond, J.Q.; Fisher, R.N.; Robertson, J.M. Natural and anthropogenic landscape factors shape functional connectivity of an ecological specialist in urban Southern California. Mol. Ecol. 2022, 31, 5214–5230. [Google Scholar] [CrossRef]
- Dandois, J.P.; Ellis, E.C. High spatial resolution three-dimensional mapping of vegetation spectral dynamics using computer vision. Remote Sens. Environ. 2013, 136, 259–276. [Google Scholar] [CrossRef] [Green Version]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. Lidar remote sensing for ecosystem studies. BioScience 2002, 52, 19–30. [Google Scholar] [CrossRef]
- Vasey, N.; Mogilewsky, M.; Schatz, G.E. Infant nest and stash sites of variegated lemurs (Varecia rubra): The extended phenotype. Am. J. Primatol. 2018, 80, e22911. [Google Scholar] [CrossRef]
- Markolf, M.; Kappeler, P.M. Phylogeographic analysis of the true lemurs (genus Eulemur) underlines the role of river catchments for the evolution of micro-endemism in Madagascar. Front. Zool. 2013, 10, 70. [Google Scholar] [CrossRef] [Green Version]
- Cullingham, C.I.; Kyle, C.J.; Pond, B.A.; Rees, E.E.; White, B.N. Differential permeability of rivers to raccoon gene flow corresponds to rabies incidence in Ontario, Canada. Mol. Ecol. 2009, 18, 43–53. [Google Scholar] [CrossRef]
- Godinho, M.B.D.C.; Da Silva, F.R. The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Sci. Rep. 2018, 8, 3427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieilledent, G.; Grinand, C.; Rakotomalala, F.A.; Ranaivosoa, R.; Rakotoarijaona, J.R.; Allnutt, T.F.; Achard, F. Combining global tree cover loss data with historical national forest cover maps to look at six decades of deforestation and forest fragmentation in Madagascar. Biol. Conserv. 2018, 222, 189–197. [Google Scholar] [CrossRef]
- Green, G.M.; Sussman, R.W. Deforestation history of the eastern rain forests of Madagascar from satellite images. Science 1990, 248, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Grinand, C.; Rakotomalala, F.; Gond, V.; Vaudry, R.; Bernoux, M.; Vieilledent, G. Estimating deforestation in tropical humid and dry forests in Madagascar from 2000 to 2010 using multi-date Landsat satellite images and the random forests classifier. Remote Sens. Environ. 2013, 139, 68–80. [Google Scholar] [CrossRef]
- Harper, G.J.; Steininger, M.K.; Tucker, C.J.; Juhn, D.; Hawkins, F. Fifty years of deforestation and forest fragmentation in Madagascar. Environ. Conserv. 2007, 34, 325–333. [Google Scholar] [CrossRef]
- Ramiadantsoa, T.; Ovaskainen, O.; Rybicki, J.; Hanski, I. Large-scale habitat corridors for biodiversity conservation: A forest corridor in Madagascar. PLoS ONE 2015, 10, e0132126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaudrot, L.; Kamilar, J.M.; Marshall, A.J.; Reed, K.E. African primate assemblages exhibit a latitudinal gradient in dispersal limitation. Int. J. Primatol. 2014, 35, 1088–1104. [Google Scholar] [CrossRef]
- Sales, L.P.; Ribeiro, B.R.; Pires, M.M.; Chapman, C.A.; Loyola, R. Recalculating route: Dispersal constraints will drive the redistribution of Amazon primates in the Anthropocene. Ecography 2019, 42, 1789–1801. [Google Scholar] [CrossRef] [Green Version]
- Friedl, M.A.; Brodley, C.E. Decision Tree Classification of Land Cover from Remotely Sensed Data. Remote Sens. Environ. 1997, 61, 399–409. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-Century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [Green Version]
- Small, C. The Landsat ETM+ spectral mixing space. Remote Sens. Environ. 2004, 93, 1–17. [Google Scholar] [CrossRef]
- Sousa, D.; Small, C. Global cross-calibration of Landsat spectral mixture models. Remote Sens. Environ. 2017, 192, 139–149. [Google Scholar] [CrossRef] [Green Version]








| Layer | k | Avg. Rank | ||
|---|---|---|---|---|
| TRI † | 4 | 2.278 | 0.285 | 63.34 |
| Canopy Height | 4 | 2.539 | 0.097 | 25.20 |
| TPI ‡ | 4 | 3.671 | 0.053 | 9.42 |
| 1990 Forest Cover | 3 | 5.436 | 0.104 | 0.38 |
| 2016 Forest Cover | 3 | 5.614 | 0.105 | 0.96 |
| Altitude | 4 | 6.064 | 0.027 | 0.30 |
| Rivers | 3 | 6.125 | 0.082 | 0.01 |
| Fire Density | 4 | 6.199 | 0.025 | 0.39 |
| Distance | 2 | 7.078 | 0.223 | 0.00 |
| Surface | Log-Likelihood | AICc | Equation | Shape | Maximum |
|---|---|---|---|---|---|
| Altitude | 1798.387 | −3588.073 | Reverse Monomolecular | 7.381 | 5.192 |
| Canopy Height | 1800.029 | −3591.357 | Inverse Ricker | 4.857 | 364.762 |
| Fire Density | 1798.126 | −3587.551 | Inverse-Reverse Ricker | 4.972 | 2770.590 |
| TPI ‡ | 1799.192 | −3589.682 | Inverse-Reverse Ricker | 3.624 | 2568.438 |
| TRI § | 1801.033 | −3593.364 | Inverse Ricker | 2.606 | 3490.174 |
| Distance | 1797.957 | −3591.710 | - | - | - |
| Surface | Log-Likelihood | AICc | Feature 1: Resistance | Feature 1 | Feature 2: Resistance | Feature 2 |
|---|---|---|---|---|---|---|
| 1990 Forest Cover | 1798.493 | −3590.572 | 12.740 | Matrix | 1 | Forest Cover |
| 2016 Forest Cover | 1798.511 | −3588.066 | 3.185 | Matrix | 1 | Forest Cover |
| Rivers | 1798.280 | −3590.607 | 1 | Non-rivers | 17.175 | Rivers |
| Distance | 1797.957 | −3591.710 | - | - | - | - |
| Layer | k | Avg. Rank | ||
|---|---|---|---|---|
| Combination C | 7 | 3.045 | 0.007 | 12.15 |
| TRI † | 4 | 3.104 | 0.375 | 30.65 |
| Combination D | 7 | 3.458 | 0.007 | 18.56 |
| Canopy Height | 4 | 4.347 | 0.161 | 17.53 |
| Combination A | 10 | 4.450 | <0.001 | 5.02 |
| Combination B | 7 | 4.568 | 0.002 | 8.84 |
| TPI ‡ | 4 | 5.515 | 0.087 | 7.23 |
| Distance | 2 | 7.513 | 0.361 | 0.02 |
| Surface | Log Likelihood | AICc | Canopy Height Trans | Canopy Height Shape | Canopy Height Max | TPI ‡ Trans | TPI ‡ Shape | TPI ‡ Max | TRI § Trans | TRI § Shape | TRI § Max |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Comb. A | 1799.883 | −3575.451 | Inverse Ricker | 4.27 | 9250.48 | Inverse-Reverse Ricker | 3.08 | 14.23 | Inverse Ricker | 2.43 | 4422.37 |
| Comb. B | 1800.073 | −3584.243 | Inverse Ricker | 4.82 | 3974.92 | Ricker | 0.76 | 5483.57 | - | - | - |
| Comb. C | 1801.071 | −3582.323 | Distance | 2.77 | 5939.49 | - | - | - | Inverse Ricker | 2.57 | 9837.73 |
| Comb. D | 1801.025 | −3586.255 | - | - | - | Inverse Monomolecular | 0.59 | 2957.12 | Inverse Ricker | 2.57 | 8347.75 |
| Distance | 1797.957 | −3584.071 | - | - | - | - | - | - | - | - | - |
| Model | K | Log-Likelihood | AIC |
|---|---|---|---|
| IVI † | 1 | 722.129 | −1434.258 |
| NDVI ‡ | 1 | 721.557 | −1433.114 |
| IBD § | 1 | 721.248 | −1434.496 |
| Productivity | 3 | 721.063 | −1428.126 |
| Canopy Height | 1 | 719.966 | −1429.932 |
| ENS †† | 1 | 719.835 | −1429.671 |
| Stem Density | 1 | 719.619 | −1429.238 |
| Topography | 1 | 719.373 | −1428.747 |
| Basal Area | 1 | 719.137 | −1428.275 |
| Structure | 3 | 717.375 | −1420.750 |
| Model | K | log-likelihood | AIC |
|---|---|---|---|
| NDVI + Canopy Height Resistance | 2 | 758.784 | −1505.567 |
| Canopy Height Resistance | 1 | 758.069 | −1506.137 |
| NDVI + IVI + Canopy Height Resistance | 3 | 757.557 | −1501.114 |
| IVI + Canopy Height Resistance | 2 | 757.280 | −1502.560 |
| NDVI + Canopy Height Resistance + TRI Resistance | 3 | 756.445 | −1498.889 |
| Canopy Height Resistance + TRI Resistance | 2 | 755.811 | −1499.621 |
| NDVI + IVI + Canopy Height Resistance + TRI Resistance | 4 | 755.188 | −1494.377 |
| IVI + Canopy Height Resistance + TRI Resistance | 3 | 754.942 | −1495.884 |
| NDVI + IVI + TRI Resistance | 3 | 730.210 | −1446.420 |
| NDVI + TRI Resistance | 2 | 730.004 | −1448.008 |
| IVI + TRI Resistance | 2 | 729.988 | −1447.977 |
| TRI Resistance | 1 | 729.764 | −1449.529 |
| NDVI + IVI | 2 | 722.366 | −1432.731 |
| IVI † | 1 | 722.129 | −1434.258 |
| NDVI ‡ | 1 | 721.557 | −1433.114 |
| IBD § | 1 | 721.248 | −1434.496 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancini, A.N.; Chandrashekar, A.; Lahitsara, J.P.; Ogbeta, D.G.; Rajaonarivelo, J.A.; Ranaivorazo, N.R.; Rasoazanakolona, J.; Safwat, M.; Solo, J.; Razafindraibe, J.G.; et al. Terrain Ruggedness and Canopy Height Predict Short-Range Dispersal in the Critically Endangered Black-and-White Ruffed Lemur. Genes 2023, 14, 746. https://doi.org/10.3390/genes14030746
Mancini AN, Chandrashekar A, Lahitsara JP, Ogbeta DG, Rajaonarivelo JA, Ranaivorazo NR, Rasoazanakolona J, Safwat M, Solo J, Razafindraibe JG, et al. Terrain Ruggedness and Canopy Height Predict Short-Range Dispersal in the Critically Endangered Black-and-White Ruffed Lemur. Genes. 2023; 14(3):746. https://doi.org/10.3390/genes14030746
Chicago/Turabian StyleMancini, Amanda N., Aparna Chandrashekar, Jean Pierre Lahitsara, Daisy Gold Ogbeta, Jeanne Arline Rajaonarivelo, Ndimbintsoa Rojoarinjaka Ranaivorazo, Joseane Rasoazanakolona, Mayar Safwat, Justin Solo, Jean Guy Razafindraibe, and et al. 2023. "Terrain Ruggedness and Canopy Height Predict Short-Range Dispersal in the Critically Endangered Black-and-White Ruffed Lemur" Genes 14, no. 3: 746. https://doi.org/10.3390/genes14030746