Smoking Suppresses the Therapeutic Potential of Adipose Stem Cells in Crohn’s Disease Patients through Epigenetic Changes
Abstract
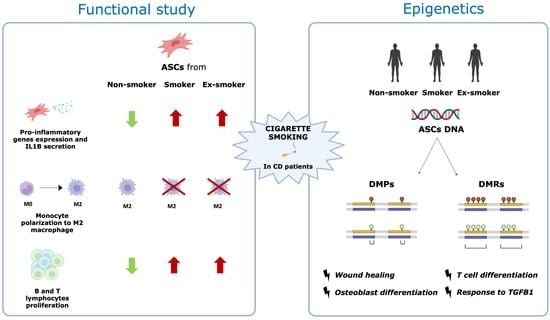
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Adipose Tissue-Derived Stem Cells
2.3. Adipose Stem Cell Immunophenotyping
2.4. THP-1, JURKAT and MEC-1 Cell Culture
2.5. Stimulation of THP-1 Cells
2.6. RNA Extraction
2.7. Real-Time Quantitative PCR
2.8. T- and B-Cell Proliferation Assay
2.9. Analysis of Differentially Methylated Loci
2.10. Functional Network Analysis and Visualization
2.11. Statistical Analysis
3. Results
3.1. Smoking Disrupts the Immune Regulatory Properties of Adipose-Derived Stem Cells from Patients with Crohn’s Disease
3.2. Smoking Induces Changes in the Methylation Pattern of Adipose-Derived Stem Cells from Patients with Crohn’s Disease
3.3. Smoking-Associated Differentially Methylated Positions in Adipose-Derived Stem Cells from Patients with Crohn’s Disease Related to Regenerative Properties
3.4. Smoking Affects the Regenerative and Anti-Inflammatory Properties of ASCs, Which Endure after Smoking Cessation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, S.S.M.; Hart, A. Aetiology and Clinical Features of Crohn’s Disease. In Crohn’s Disease: Current Concepts; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 1–10. ISBN 9783319019130. [Google Scholar]
- Volk, N.; Siegel, C.A. Defining Failure of Medical Therapy for Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2019, 25, 74–77. [Google Scholar] [CrossRef]
- Reynolds, I.S.; Doogan, K.L.; Ryan, É.J.; Hechtl, D.; Lecot, F.P.; Arya, S.; Martin, S.T. Surgical Strategies to Reduce Postoperative Recurrence of Crohn’s Disease After Ileocolic Resection. Front. Surg. 2021, 8, 804137. [Google Scholar] [CrossRef] [PubMed]
- Alqasrawi, D.; Abdelli, L.S.; Naser, S.A. Mystery Solved: Why Smoke Extract Worsens Disease in Smokers with Crohn’s Disease and Not Ulcerative Colitis? Gut Map! Microorganisms 2020, 8, 666. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yao, Q.; Chen, W.; Gao, F.; Li, P.; Wu, J.; Yu, J.; Cao, H. Stem Cell Therapy for Crohn’s Disease: Systematic Review and Meta-Analysis of Preclinical and Clinical Studies. Stem Cell Res. Ther. 2021, 12, 463. [Google Scholar] [CrossRef] [PubMed]
- Panés, J.; García-Olmo, D.; Van Assche, G.; Colombel, J.F.; Reinisch, W.; Baumgart, D.C.; Dignass, A.; Nachury, M.; Ferrante, M.; Kazemi-Shirazi, L.; et al. Long-Term Efficacy and Safety of Stem Cell Therapy (Cx601) for Complex Perianal Fistulas in Patients With Crohn’s Disease. Gastroenterology 2018, 154, 1334–1342.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Ghadban, S.; Bunnell, B.A. Adipose Tissue-Derived Stem Cells: Immunomodulatory Effects and Therapeutic Potential. Physiology 2020, 35, 125–133. [Google Scholar] [CrossRef]
- Serena, C.; Keiran, N.; Madeira, A.; Maymó-Masip, E.; Ejarque, M.; Terrón-Puig, M.; Espin, E.; Martí, M.; Borruel, N.; Guarner, F.; et al. Crohn’s Disease Disturbs the Immune Properties of Human Adipose-Derived Stem Cells Related to Inflammasome Activation. Stem. Cell Rep. 2017, 9, 1109–1123. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; El-Sabbagh, A.S.; Lukas, B.E.; Tanneberger, S.J.; Jiang, Y. Adipose Stem Cells in Obesity: Challenges and Opportunities. Biosci. Rep. 2020, 40, BSR20194076. [Google Scholar] [CrossRef]
- Park, J.S.; Park, G.; Hong, H.S. Age Affects the Paracrine Activity and Differentiation Potential of Human Adipose-Derived Stem Cells. Mol. Med. Rep. 2021, 23, 160. [Google Scholar] [CrossRef]
- Harrell, C.R.; Djonov, V.; Volarevic, V. The Effects of Cigarette Smoking and Nicotine on the Therapeutic Potential of Mesenchymal Stem Cells. Histol. Histopathol. 2022, 37, 93–100. [Google Scholar]
- Alessio, N.; Acar, M.B.; Demirsoy, I.H.; Squillaro, T.; Siniscalco, D.; Di Bernardo, G.; Peluso, G.; Özcan, S.; Galderisi, U. Obesity Is Associated with Senescence of Mesenchymal Stromal Cells Derived from Bone Marrow, Subcutaneous and Visceral Fat of Young Mice. Aging 2020, 12, 12609–12621. [Google Scholar] [CrossRef] [PubMed]
- Saheli, M.; Khoramipour, K.; Vosough, M.; Piryaei, A.; Rahmati, M.; Suzuki, K. Athletes’ Mesenchymal Stem Cells Could Be the Best Choice for Cell Therapy in Omicron-Infected Patients. Cells 2022, 11, 1926. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Chen, Q.; Xie, M. Smoking Increases the Risk of Infectious Diseases: A Narrative Review. Tob. Induc. Dis. 2020, 18, 60. [Google Scholar] [CrossRef]
- Umnuaypornlert, A.; Kanchanasurakit, S.; Lucero-Prisno, D.E.; Saokaew, S. Smoking and Risk of Negative Outcomes among COVID-19 Patients: A Systematic Review and Meta-Analysis. Tob. Induc. Dis. 2021, 19, 9. [Google Scholar] [CrossRef]
- Liu, C.; Marioni, R.E.; Hedman, A.K.; Pfeiffer, L.; Tsai, P.C.; Reynolds, L.M.; Just, A.C.; Duan, Q.; Boer, C.G.; Tanaka, T.; et al. A DNA Methylation Biomarker of Alcohol Consumption. Mol. Psychiatry 2018, 23, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Tebyaniyan, H.; Khayatan, D. The Role of Epigenetic in Dental and Oral Regenerative Medicine by Different Types of Dental Stem Cells: A Comprehensive Overview. Stem. Cells Int. 2022, 2022, 5304860. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.M.; Carballosa, C.M.; Cheung, H.S. Concise Review: The Deleterious Effects of Cigarette Smoking and Nicotine Usage and Mesenchymal Stem Cell Function and Implications for Cell-Based Therapies. Stem. Cells Transl. Med. 2017, 6, 1815–1821. [Google Scholar] [CrossRef]
- Best, W.R.; Becktel, J.M.; Singleton, J.W.; Kern, F. Development of a Crohn’s Disease Activity Index: National Cooperative Crohn’s Disease Study. Gastroenterology 1976, 70, 439–444. [Google Scholar] [CrossRef]
- Van Assche, G.; Dignass, A.; Reinisch, W.; van der Woude, C.J.; Sturm, A.; De Vos, M.; Guslandi, M.; Oldenburg, B.; Dotan, I.; Marteau, P.; et al. The Second European Evidence-Based Consensus on the Diagnosis and Management of Crohn’s Disease: Special Situations. J. Crohns Colitis 2010, 4, 63–101. [Google Scholar] [CrossRef] [Green Version]
- Dubois, S.G.; Floyd, E.Z.; Zvonic, S.; Kilroy, G.; Wu, X.; Carling, S.; Di, Y.; Halvorsen, C.; Ravussin, E.; Gimble, J.M. Isolation of Human Adipose-Derived Stem Cells from Biopsies and Liposuction Specimens. Mesenchimal Stem. Cells 2008, 5, 69–79. [Google Scholar] [CrossRef]
- Pachón-Peña, G.; Serena, C.; Ejarque, M.; Petriz, J.; Duran, X.; Oliva-Olivera, W.; Simó, R.; Tinahones, F.J.; Fernández-Veledo, S.; Vendrell, J. Obesity Determines the Immunophenotypic Profile and Functional Characteristics of Human Mesenchymal Stem Cells From Adipose Tissue. Stem. Cells Transl. Med. 2016, 5, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Serena, C.; Millan, M.; Ejarque, M.; Saera-Vila, A.; Maymó-Masip, E.; Núñez-Roa, C.; Monfort-Ferré, D.; Terrón-Puig, M.; Bautista, M.; Menacho, M.; et al. Adipose Stem Cells from Patients with Crohn’s Disease Show a Distinctive DNA Methylation Pattern. Clin. Epigenet. 2020, 12, 53. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Müller, F.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Comprehensive Analysis of DNA Methylation Data with RnBeads. Nat. Methods 2014, 11, 1138–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, F.; Scherer, M.; Assenov, Y.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. RnBeads 2.0: Comprehensive Analysis of DNA Methylation Data. Genome Biol. 2019, 20, 55. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.L.; Baggerly, K.A.; Bengtsson, H.; Ritchie, M.E.; Hansen, K.D. Illuminaio: An Open Source IDAT Parsing Tool for Illumina Microarrays. F1000Res 2013, 2, 264. [Google Scholar] [CrossRef] [Green Version]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A Beta-Mixture Quantile Normalization Method for Correcting Probe Design Bias in Illumina Infinium 450 k DNA Methylation Data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Jaffe, A.E.; Murakami, P.; Lee, H.; Leek, J.T.; Fallin, M.D.; Feinberg, A.P.; Irizarry, R.A. Bump Hunting to Identify Differentially Methylated Regions in Epigenetic Epidemiology Studies. Int. J. Epidemiol. 2012, 41, 200–209. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Benjaminit, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Wickham, H. Ggplot2; Springer: Houston, TX, USA, 2009. [Google Scholar]
- Yamamoto, T.; Keighley, M.R.B. Smoking and Disease Recurrence after Operation for Crohn’s Disease. Br. J. Surg. 2000, 87, 398. [Google Scholar] [CrossRef]
- Nguyen, B.; Alpagot, T.; Oh, H.; Ojcius, D.; Xiao, N. Comparison of the Effect of Cigarette Smoke on Mesenchymal Stem Cells and Dental Stem Cells. Am. J. Physiol. Cell Physiol. 2021, 320, C175–C181. [Google Scholar] [CrossRef] [PubMed]
- Barwinska, D.; Traktuev, D.O.; Merfeld-Clauss, S.; Cook, T.G.; Lu, H.; Petrache, I.; March, K.L. Cigarette Smoking Impairs Adipose Stromal Cell Vasculogenic Activity and Abrogates Potency to Ameliorate Ischemia. Stem. Cells 2018, 36, 856–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiori, A.; Uhlig, S.; Klüter, H.; Bieback, K. Human Adipose Tissue-Derived Mesenchymal Stromal Cells Inhibit CD4+ T Cell Proliferation and Induce Regulatory T Cells as Well as CD127 Expression on CD4+CD25+ T Cells. Cells 2021, 10, 58. [Google Scholar] [CrossRef]
- Heo, J.S.; Choi, Y.; Kim, H.O. Adipose-Derived Mesenchymal Stem Cells Promote M2 Macrophage Phenotype through Exosomes. Stem. Cells Int. 2019, 2019, 7921760. [Google Scholar] [CrossRef]
- Afarideh, M.; Thaler, R.; Khani, F.; Tang, H.; Jordan, K.L.; Conley, S.M.; Saadiq, I.M.; Obeidat, Y.; Pawar, A.S.; Eirin, A.; et al. Global Epigenetic Alterations of Mesenchymal Stem Cells in Obesity: The Role of Vitamin C Reprogramming. Epigenetics 2021, 16, 705–717. [Google Scholar] [CrossRef]
- Guo, X.; Schaudinn, C.; Blume-Peytavi, U.; Vogt, A.; Rancan, F. Effects of Adipose-Derived Stem Cells and Their Conditioned Medium in a Human Ex Vivo Wound Model. Cells 2022, 11, 1198. [Google Scholar] [CrossRef]
- Xu, X.; Zheng, L.; Yuan, Q.; Zhen, G.; Crane, J.L.; Zhou, X.; Cao, X. Transforming Growth Factor-β in Stem Cells and Tissue Homeostasis. Bone Res. 2018, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Vieujean, S.; Caron, B.; Haghnejad, V.; Jouzeau, J.Y.; Netter, P.; Heba, A.C.; Ndiaye, N.C.; Moulin, D.; Barreto, G.; Danese, S.; et al. Impact of the Exposome on the Epigenome in Inflammatory Bowel Disease Patients and Animal Models. Int. J. Mol. Sci. 2022, 23, 7611. [Google Scholar] [CrossRef]
- Dhar, G.A.; Saha, S.; Mitra, P.; Nag Chaudhuri, R. DNA Methylation and Regulation of Gene Expression: Guardian of Our Health. Nucleus 2021, 64, 259–270. [Google Scholar] [CrossRef]
- Garrett-Sinha, L.A. Review of Ets1 Structure, Function, and Roles in Immunity. Cell. Mol. Life Sci. 2013, 70, 3375–3390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gluck, C.; Glathar, A.; Tsompana, M.; Nowak, N.; Garrett-Sinha, L.A.; Buck, M.J.; Sinha, S. Molecular Dissection of the Oncogenic Role of ETS1 in the Mesenchymal Subtypes of Head and Neck Squamous Cell Carcinoma. PLoS Genet. 2019, 15, e1008250. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Gao, H.; Chang, Y.L.; Wu, X.; Lin, R.; Li, G.; Lin, J.; Lu, H.; Chen, H.; Li, Z.; et al. ETS-1 Facilitates Th1 Cell-Mediated Mucosal Inflammation in Inflammatory Bowel Diseases through Upregulating CIRBP: Role of ETS-1 and CIRBP in IBD. J. Autoimmun. 2022, 132, 102872. [Google Scholar] [CrossRef] [PubMed]
- Wahl, E.A.; Schenck, T.L.; Machens, H.G.; Egaña, J.T. Acute Stimulation of Mesenchymal Stem Cells with Cigarette Smoke Extract Affects Their Migration, Differentiation, and Paracrine Potential. Sci. Rep. 2016, 6, 22957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gersemann, M.; Stange, E.F.; Wehkamp, J. From Intestinal Stem Cells to Inflammatory Bowel Diseases. World J. Gastroenterol. 2011, 17, 3198–3203. [Google Scholar] [CrossRef]
- Egging, D.F.; Peeters, A.C.T.M.; Grebenchtchikov, N.; Geurts-Moespot, A.; Sweep, C.G.J.; Den Heijer, M.; Schalkwijk, J. Identification and Characterization of Multiple Species of Tenascin-X in Human Serum. FEBS J. 2007, 274, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Barrow, T.M.; Klett, H.; Toth, R.; Böhm, J.; Gigic, B.; Habermann, N.; Scherer, D.; Schrotz-King, P.; Skender, S.; Abbenhardt-Martin, C.; et al. Smoking Is Associated with Hypermethylation of the APC 1A Promoter in Colorectal Cancer: The ColoCare Study. J. Pathol. 2017, 243, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Marino, R.; Moresco, A.; Perez Garrido, N.; Ramirez, P.; Belgorosky, A. Congenital Adrenal Hyperplasia and Ehlers-Danlos Syndrome. Front. Endocrinol. 2022, 13, 227. [Google Scholar] [CrossRef]
- Green, C.; Ghali, N.; Akilapa, R.; Angwin, C.; Baker, D.; Bartlett, M.; Bowen, J.; Brady, A.F.; Brock, J.; Chamberlain, E.; et al. Classical-like Ehlers-Danlos Syndrome: A Clinical Description of 20 Newly Identified Individuals with Evidence of Tissue Fragility. Genet. Med. 2020, 22, 1576–1582. [Google Scholar] [CrossRef]
- Suau, R.; Pardina, E.; Domènech, E.; Lorén, V.; Manyé, J. The Complex Relationship Between Microbiota, Immune Response and Creeping Fat in Crohn’s Disease. J. Crohns Colitis. 2022, 16, 472–489. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boronat-Toscano, A.; Vañó, I.; Monfort-Ferré, D.; Menacho, M.; Valldosera, G.; Caro, A.; Espina, B.; Mañas, M.J.; Marti, M.; Espin, E.; et al. Smoking Suppresses the Therapeutic Potential of Adipose Stem Cells in Crohn’s Disease Patients through Epigenetic Changes. Cells 2023, 12, 1021. https://doi.org/10.3390/cells12071021
Boronat-Toscano A, Vañó I, Monfort-Ferré D, Menacho M, Valldosera G, Caro A, Espina B, Mañas MJ, Marti M, Espin E, et al. Smoking Suppresses the Therapeutic Potential of Adipose Stem Cells in Crohn’s Disease Patients through Epigenetic Changes. Cells. 2023; 12(7):1021. https://doi.org/10.3390/cells12071021
Chicago/Turabian StyleBoronat-Toscano, Albert, Irene Vañó, Diandra Monfort-Ferré, Margarita Menacho, Gemma Valldosera, Aleidis Caro, Beatriz Espina, Maria José Mañas, Marc Marti, Eloy Espin, and et al. 2023. "Smoking Suppresses the Therapeutic Potential of Adipose Stem Cells in Crohn’s Disease Patients through Epigenetic Changes" Cells 12, no. 7: 1021. https://doi.org/10.3390/cells12071021